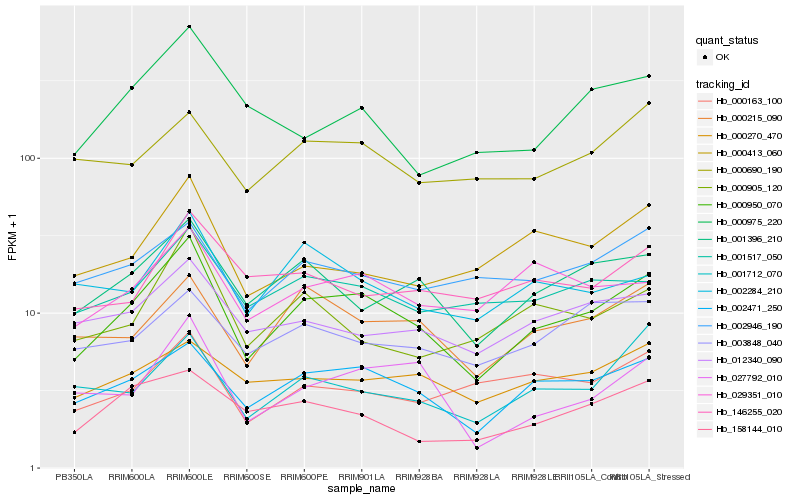
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000950_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645259 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002471_250 |
0.1002561002 |
- |
- |
Tapt1/CMV receptor, putative isoform 2 [Theobroma cacao] |
| 3 |
Hb_001396_210 |
0.1266421886 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000215_090 |
0.136047803 |
- |
- |
cytochrome b5 domain-containing family protein [Populus trichocarpa] |
| 5 |
Hb_027792_010 |
0.1396621138 |
- |
- |
- |
| 6 |
Hb_001517_050 |
0.1428563892 |
transcription factor |
TF Family: BBR-BPC |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_158144_010 |
0.1473309937 |
- |
- |
- |
| 8 |
Hb_012340_090 |
0.1474979051 |
- |
- |
PREDICTED: endonuclease III homolog 1, chloroplastic isoform X1 [Populus euphratica] |
| 9 |
Hb_001712_070 |
0.1483441646 |
- |
- |
PREDICTED: HD domain-containing protein 2 [Jatropha curcas] |
| 10 |
Hb_029351_010 |
0.1496554365 |
- |
- |
PREDICTED: HD domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002284_210 |
0.1509066334 |
- |
- |
PREDICTED: protein canopy-1 [Jatropha curcas] |
| 12 |
Hb_000905_120 |
0.1523766593 |
- |
- |
PREDICTED: uncharacterized protein LOC105628677 [Jatropha curcas] |
| 13 |
Hb_002946_190 |
0.1526764093 |
- |
- |
PREDICTED: nudix hydrolase 26, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000270_470 |
0.1528761767 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: pentatricopeptide repeat-containing protein At1g74900, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000413_060 |
0.1538402609 |
- |
- |
PREDICTED: DAG protein, chloroplastic [Populus euphratica] |
| 16 |
Hb_000163_100 |
0.1540631448 |
- |
- |
PREDICTED: uncharacterized protein LOC105642537 [Jatropha curcas] |
| 17 |
Hb_000975_220 |
0.1548580496 |
- |
- |
Histone H2B [Medicago truncatula] |
| 18 |
Hb_003848_040 |
0.1567234663 |
- |
- |
PREDICTED: phosphatidylinositol-glycan biosynthesis class X protein isoform X1 [Jatropha curcas] |
| 19 |
Hb_000690_190 |
0.1569842641 |
- |
- |
40S ribosomal protein S7, putative [Ricinus communis] |
| 20 |
Hb_146255_020 |
0.1573207197 |
- |
- |
PREDICTED: chromatin structure-remodeling complex protein BSH [Jatropha curcas] |