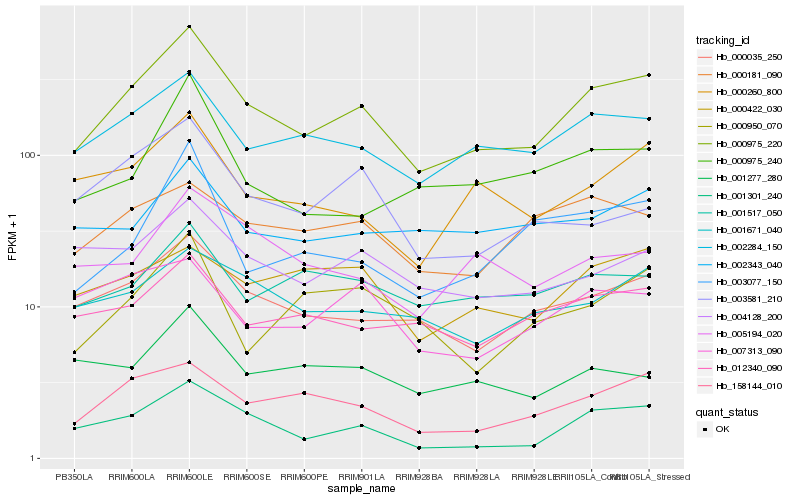
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000975_220 |
0.0 |
- |
- |
Histone H2B [Medicago truncatula] |
| 2 |
Hb_001301_240 |
0.1013842419 |
- |
- |
- |
| 3 |
Hb_000035_250 |
0.1142456335 |
- |
- |
PREDICTED: uncharacterized protein LOC105637544 [Jatropha curcas] |
| 4 |
Hb_002284_150 |
0.1161341036 |
- |
- |
PREDICTED: uncharacterized protein LOC105635198 [Jatropha curcas] |
| 5 |
Hb_158144_010 |
0.1208924965 |
- |
- |
- |
| 6 |
Hb_000260_800 |
0.1369874255 |
- |
- |
PREDICTED: probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 [Populus euphratica] |
| 7 |
Hb_001671_040 |
0.1416075658 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS31-like isoform X1 [Populus euphratica] |
| 8 |
Hb_012340_090 |
0.1426071771 |
- |
- |
PREDICTED: endonuclease III homolog 1, chloroplastic isoform X1 [Populus euphratica] |
| 9 |
Hb_004128_200 |
0.1447357552 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |
| 10 |
Hb_001517_050 |
0.1494665709 |
transcription factor |
TF Family: BBR-BPC |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003077_150 |
0.149500856 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 12 |
Hb_000975_240 |
0.1515405846 |
- |
- |
hypothetical protein JCGZ_22006 [Jatropha curcas] |
| 13 |
Hb_002343_040 |
0.1525118277 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_005194_020 |
0.1535413808 |
- |
- |
PREDICTED: UPF0553 protein-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000181_090 |
0.1541789334 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 16 |
Hb_000950_070 |
0.1548580496 |
- |
- |
PREDICTED: uncharacterized protein LOC105645259 isoform X2 [Jatropha curcas] |
| 17 |
Hb_007313_090 |
0.1565103423 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000422_030 |
0.1582926025 |
- |
- |
PREDICTED: uncharacterized protein LOC105643943 [Jatropha curcas] |
| 19 |
Hb_003581_210 |
0.1591706643 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-binding protein WHY1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001277_280 |
0.1593751738 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |