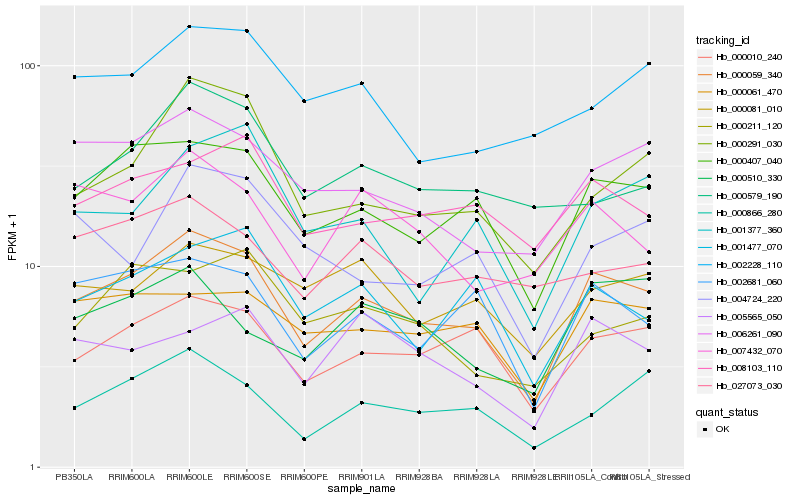
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000059_340 |
0.0 |
transcription factor |
TF Family: SOH1 |
uncharacterized protein LOC100527879 [Glycine max] |
| 2 |
Hb_000407_040 |
0.0846636211 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_000010_240 |
0.104816043 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 4 |
Hb_002681_060 |
0.1140266012 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 5 |
Hb_006261_090 |
0.1202471062 |
- |
- |
PREDICTED: exosome complex exonuclease RRP46 homolog [Jatropha curcas] |
| 6 |
Hb_000061_470 |
0.1222307485 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: DNA annealing helicase and endonuclease ZRANB3 [Jatropha curcas] |
| 7 |
Hb_000866_280 |
0.1265040599 |
- |
- |
PREDICTED: seven transmembrane domain-containing tyrosine-protein kinase 1-like [Jatropha curcas] |
| 8 |
Hb_008103_110 |
0.1295452185 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_000291_030 |
0.1304250758 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Eucalyptus grandis] |
| 10 |
Hb_001477_070 |
0.1308346316 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_007432_070 |
0.1355008568 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 12 |
Hb_000081_010 |
0.1365209675 |
- |
- |
hypothetical protein B456_012G056000 [Gossypium raimondii] |
| 13 |
Hb_001377_360 |
0.1398089406 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18 [Jatropha curcas] |
| 14 |
Hb_005565_050 |
0.1410297428 |
- |
- |
hypothetical protein CICLE_v10010292mg [Citrus clementina] |
| 15 |
Hb_000510_330 |
0.1422240655 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 16 |
Hb_027073_030 |
0.1430990717 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 17 |
Hb_002228_110 |
0.1432842063 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |
| 18 |
Hb_000579_190 |
0.1443430863 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 19 |
Hb_004724_220 |
0.145609685 |
- |
- |
PREDICTED: probable calcium-binding protein CML48 [Jatropha curcas] |
| 20 |
Hb_000211_120 |
0.1463800455 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |