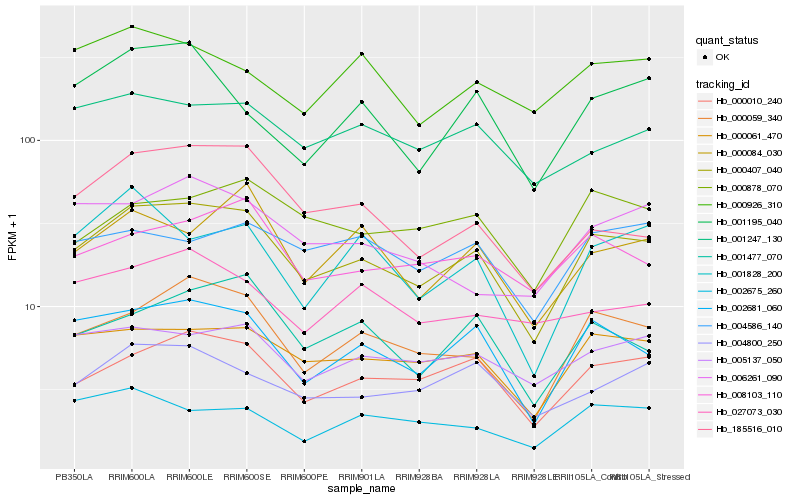
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000407_040 |
0.0 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_002681_060 |
0.08285289 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 3 |
Hb_000010_240 |
0.0844646835 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 4 |
Hb_000059_340 |
0.0846636211 |
transcription factor |
TF Family: SOH1 |
uncharacterized protein LOC100527879 [Glycine max] |
| 5 |
Hb_000061_470 |
0.0894686409 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: DNA annealing helicase and endonuclease ZRANB3 [Jatropha curcas] |
| 6 |
Hb_001477_070 |
0.1093396716 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_008103_110 |
0.1105914249 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_005137_050 |
0.1113734914 |
- |
- |
PREDICTED: nuclear pore complex protein NUP58 [Jatropha curcas] |
| 9 |
Hb_004800_250 |
0.1148854158 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A [Jatropha curcas] |
| 10 |
Hb_001247_130 |
0.1160872115 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha [Jatropha curcas] |
| 11 |
Hb_000878_070 |
0.1203888919 |
- |
- |
hypothetical protein POPTR_0016s06670g [Populus trichocarpa] |
| 12 |
Hb_000084_030 |
0.1208376923 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_185516_010 |
0.122030429 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 14 |
Hb_001195_040 |
0.1266581922 |
- |
- |
PREDICTED: proteasome subunit alpha type-1-B-like [Gossypium raimondii] |
| 15 |
Hb_002675_260 |
0.12701561 |
- |
- |
hypothetical protein JCGZ_06227 [Jatropha curcas] |
| 16 |
Hb_006261_090 |
0.1277212504 |
- |
- |
PREDICTED: exosome complex exonuclease RRP46 homolog [Jatropha curcas] |
| 17 |
Hb_001828_200 |
0.1277915329 |
- |
- |
ribosomal protein L13 [Populus trichocarpa] |
| 18 |
Hb_027073_030 |
0.1285818877 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 19 |
Hb_004586_140 |
0.1293226026 |
- |
- |
PREDICTED: uncharacterized protein LOC105630689 [Jatropha curcas] |
| 20 |
Hb_000926_310 |
0.1301246047 |
- |
- |
uncharacterized protein LOC100500241 [Glycine max] |