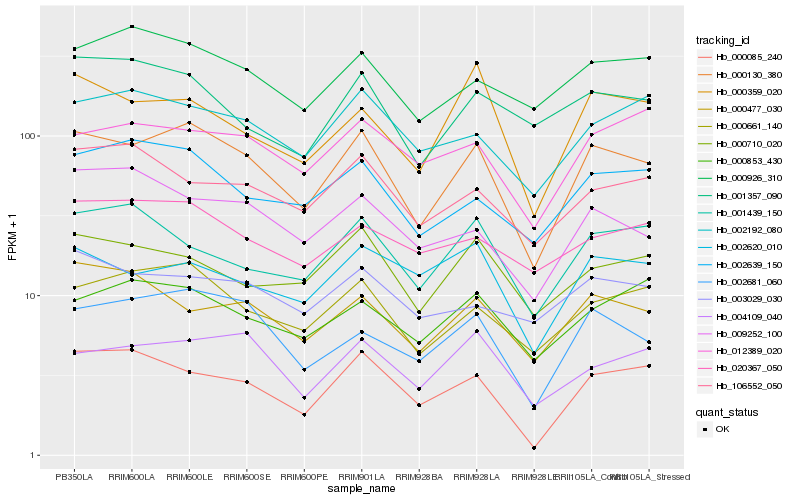
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000130_380 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635736 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000661_140 |
0.1010438659 |
- |
- |
PREDICTED: uncharacterized protein LOC105648317 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002681_060 |
0.104290329 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 4 |
Hb_004109_040 |
0.1164713494 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g53600, mitochondrial [Jatropha curcas] |
| 5 |
Hb_002639_150 |
0.1172248034 |
- |
- |
- |
| 6 |
Hb_000085_240 |
0.1173514636 |
- |
- |
fimbrin, putative [Ricinus communis] |
| 7 |
Hb_000710_020 |
0.1206146871 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MARCH2 isoform X2 [Populus euphratica] |
| 8 |
Hb_000359_020 |
0.1220521083 |
- |
- |
amine oxidase, putative [Ricinus communis] |
| 9 |
Hb_002192_080 |
0.1238516025 |
- |
- |
hypothetical protein B456_013G069200 [Gossypium raimondii] |
| 10 |
Hb_000926_310 |
0.1240765149 |
- |
- |
uncharacterized protein LOC100500241 [Glycine max] |
| 11 |
Hb_002620_010 |
0.1261725162 |
- |
- |
hypothetical protein POPTR_0004s18130g [Populus trichocarpa] |
| 12 |
Hb_000477_030 |
0.1268466214 |
- |
- |
PREDICTED: boron transporter 4-like [Jatropha curcas] |
| 13 |
Hb_001439_150 |
0.1293583457 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15340, mitochondrial [Jatropha curcas] |
| 14 |
Hb_106552_050 |
0.129408125 |
- |
- |
PREDICTED: uncharacterized protein LOC105642134 [Jatropha curcas] |
| 15 |
Hb_000853_430 |
0.129474033 |
- |
- |
PREDICTED: uncharacterized protein LOC105642597 [Jatropha curcas] |
| 16 |
Hb_012389_020 |
0.1301221576 |
- |
- |
hypothetical protein POPTR_0006s05190g [Populus trichocarpa] |
| 17 |
Hb_003029_030 |
0.1303242402 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001357_090 |
0.130558472 |
- |
- |
PREDICTED: small ubiquitin-related modifier 1 [Jatropha curcas] |
| 19 |
Hb_020367_050 |
0.1308359607 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl pyrophosphate synthase, putative [Ricinus communis] |
| 20 |
Hb_009252_100 |
0.1310510931 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |