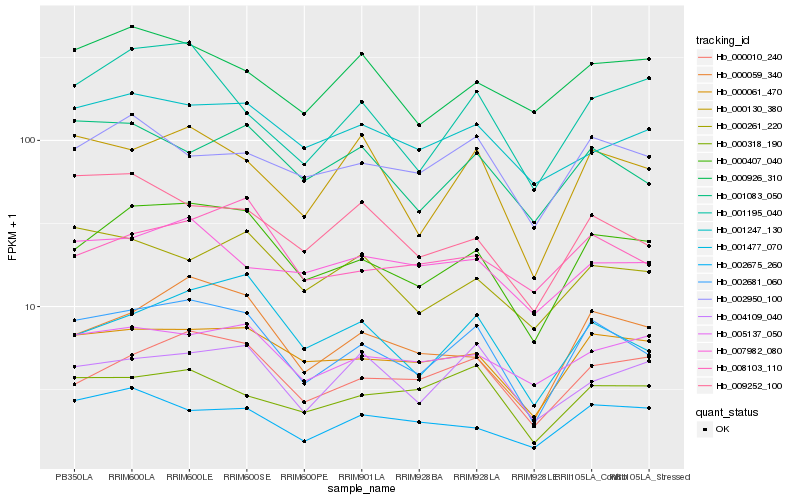
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002681_060 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 2 |
Hb_000407_040 |
0.08285289 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_000130_380 |
0.104290329 |
- |
- |
PREDICTED: uncharacterized protein LOC105635736 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000061_470 |
0.1077146 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: DNA annealing helicase and endonuclease ZRANB3 [Jatropha curcas] |
| 5 |
Hb_001477_070 |
0.1139026783 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000059_340 |
0.1140266012 |
transcription factor |
TF Family: SOH1 |
uncharacterized protein LOC100527879 [Glycine max] |
| 7 |
Hb_000010_240 |
0.1174747227 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 8 |
Hb_001083_050 |
0.1204577839 |
- |
- |
PREDICTED: V-type proton ATPase subunit d2 [Jatropha curcas] |
| 9 |
Hb_000318_190 |
0.1304678382 |
- |
- |
hypothetical protein JCGZ_04889 [Jatropha curcas] |
| 10 |
Hb_009252_100 |
0.1308265768 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 11 |
Hb_001247_130 |
0.1325558827 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha [Jatropha curcas] |
| 12 |
Hb_001195_040 |
0.1329096856 |
- |
- |
PREDICTED: proteasome subunit alpha type-1-B-like [Gossypium raimondii] |
| 13 |
Hb_004109_040 |
0.1331284639 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g53600, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000261_220 |
0.1333608192 |
- |
- |
PREDICTED: TBC1 domain family member 13 [Jatropha curcas] |
| 15 |
Hb_005137_050 |
0.1355794033 |
- |
- |
PREDICTED: nuclear pore complex protein NUP58 [Jatropha curcas] |
| 16 |
Hb_002950_100 |
0.1357727941 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S protease regulatory subunit 6B homolog [Jatropha curcas] |
| 17 |
Hb_008103_110 |
0.1359008165 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_002675_260 |
0.1370699717 |
- |
- |
hypothetical protein JCGZ_06227 [Jatropha curcas] |
| 19 |
Hb_007982_080 |
0.1390745697 |
- |
- |
PREDICTED: uncharacterized protein LOC105636531 [Jatropha curcas] |
| 20 |
Hb_000926_310 |
0.1413849142 |
- |
- |
uncharacterized protein LOC100500241 [Glycine max] |