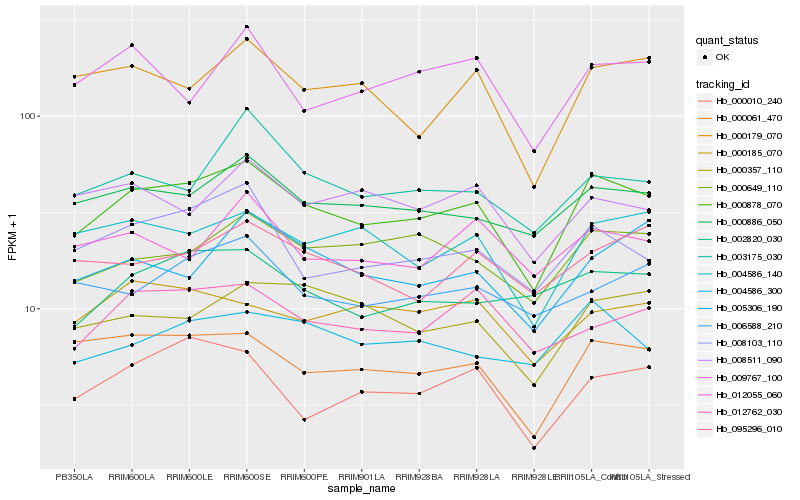
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000878_070 |
0.0 |
- |
- |
hypothetical protein POPTR_0016s06670g [Populus trichocarpa] |
| 2 |
Hb_000886_050 |
0.0884923816 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 3 |
Hb_000061_470 |
0.0921618621 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: DNA annealing helicase and endonuclease ZRANB3 [Jatropha curcas] |
| 4 |
Hb_000649_110 |
0.0947340481 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 5 |
Hb_004586_140 |
0.0948134387 |
- |
- |
PREDICTED: uncharacterized protein LOC105630689 [Jatropha curcas] |
| 6 |
Hb_008103_110 |
0.0959926036 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_004586_300 |
0.1005968628 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000179_070 |
0.1009082318 |
- |
- |
hypothetical protein CICLE_v10009411mg [Citrus clementina] |
| 9 |
Hb_000357_110 |
0.1016498523 |
- |
- |
PREDICTED: uncharacterized protein LOC105643970 isoform X2 [Jatropha curcas] |
| 10 |
Hb_012762_030 |
0.1029495527 |
- |
- |
PREDICTED: uncharacterized protein LOC105634062 [Jatropha curcas] |
| 11 |
Hb_003175_030 |
0.1047844022 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 12 |
Hb_002820_030 |
0.1059231536 |
- |
- |
PREDICTED: uncharacterized protein LOC105638922 [Jatropha curcas] |
| 13 |
Hb_005306_190 |
0.1085946426 |
- |
- |
Mitochondrial import inner membrane translocase subunit tim23, putative [Ricinus communis] |
| 14 |
Hb_000185_070 |
0.1092671473 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_095296_010 |
0.1092707204 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 16 |
Hb_012055_060 |
0.1092775625 |
- |
- |
PREDICTED: uncharacterized protein LOC105639799 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000010_240 |
0.1095773731 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 18 |
Hb_008511_090 |
0.1102095913 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 19 |
Hb_009767_100 |
0.1103694202 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 20 |
Hb_006588_210 |
0.1107948636 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Hakai [Jatropha curcas] |