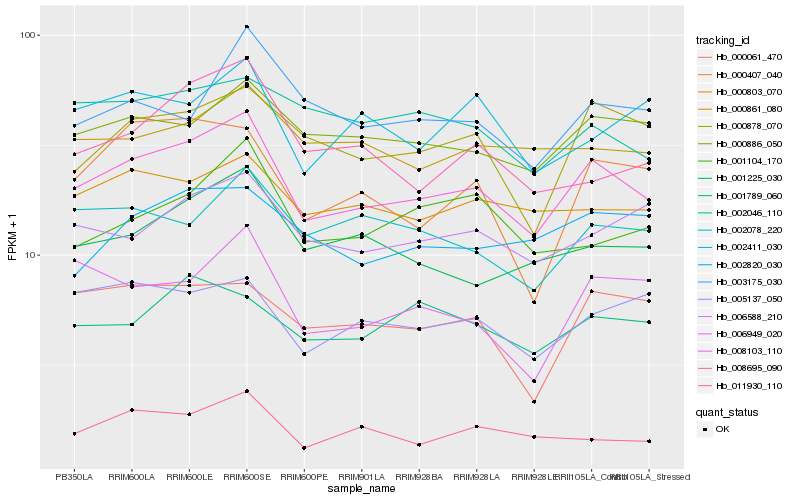
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008103_110 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_000886_050 |
0.0946398844 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 3 |
Hb_000878_070 |
0.0959926036 |
- |
- |
hypothetical protein POPTR_0016s06670g [Populus trichocarpa] |
| 4 |
Hb_001225_030 |
0.0966471653 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 5 |
Hb_000803_070 |
0.0994171275 |
- |
- |
PREDICTED: protein TRAUCO [Jatropha curcas] |
| 6 |
Hb_006588_210 |
0.1000659223 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Hakai [Jatropha curcas] |
| 7 |
Hb_005137_050 |
0.1047921936 |
- |
- |
PREDICTED: nuclear pore complex protein NUP58 [Jatropha curcas] |
| 8 |
Hb_002078_220 |
0.1049755035 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000861_080 |
0.1052777711 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 10 |
Hb_011930_110 |
0.1062476027 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g59720, mitochondrial [Jatropha curcas] |
| 11 |
Hb_003175_030 |
0.1087183178 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 12 |
Hb_008695_090 |
0.1087915024 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 13 |
Hb_001104_170 |
0.1093457759 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2c [Jatropha curcas] |
| 14 |
Hb_002820_030 |
0.1095835578 |
- |
- |
PREDICTED: uncharacterized protein LOC105638922 [Jatropha curcas] |
| 15 |
Hb_001789_060 |
0.110309631 |
- |
- |
calmodulin-binding heat-shock protein, putative [Ricinus communis] |
| 16 |
Hb_000407_040 |
0.1105914249 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_006949_020 |
0.1106900698 |
- |
- |
PREDICTED: probable 28S rRNA (cytosine-C(5))-methyltransferase isoform X1 [Jatropha curcas] |
| 18 |
Hb_000061_470 |
0.1107390799 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: DNA annealing helicase and endonuclease ZRANB3 [Jatropha curcas] |
| 19 |
Hb_002411_030 |
0.1115066471 |
transcription factor |
TF Family: NF-YC |
PREDICTED: dr1-associated corepressor homolog [Jatropha curcas] |
| 20 |
Hb_002046_110 |
0.1115807003 |
- |
- |
PREDICTED: TBC1 domain family member 15 isoform X2 [Jatropha curcas] |