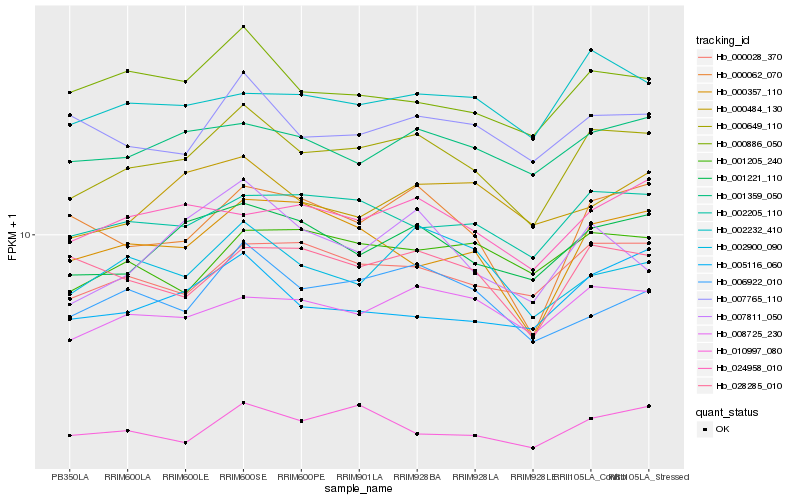
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000649_110 |
0.0 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 2 |
Hb_001221_110 |
0.0639150971 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
hypothetical protein JCGZ_11303 [Jatropha curcas] |
| 3 |
Hb_000028_370 |
0.0723149102 |
- |
- |
Guanine nucleotide-binding protein alpha-2 subunit [Glycine soja] |
| 4 |
Hb_002205_110 |
0.0732928392 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_008725_230 |
0.0744165933 |
- |
- |
PREDICTED: CTP synthase [Jatropha curcas] |
| 6 |
Hb_001359_050 |
0.0771380273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000886_050 |
0.0797534149 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 8 |
Hb_006922_010 |
0.0805175923 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 9 |
Hb_024958_010 |
0.0818646203 |
- |
- |
PREDICTED: LRR repeats and ubiquitin-like domain-containing protein At2g30105 [Jatropha curcas] |
| 10 |
Hb_010997_080 |
0.0834891132 |
transcription factor |
TF Family: mTERF |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_002232_410 |
0.0840309548 |
- |
- |
PREDICTED: L-galactose dehydrogenase [Jatropha curcas] |
| 12 |
Hb_002900_090 |
0.0842915699 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105646817 [Jatropha curcas] |
| 13 |
Hb_007765_110 |
0.0851175486 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 40-like [Jatropha curcas] |
| 14 |
Hb_000062_070 |
0.0852584749 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 15 |
Hb_028285_010 |
0.0856341766 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like [Jatropha curcas] |
| 16 |
Hb_005116_060 |
0.0859417133 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000357_110 |
0.0866588489 |
- |
- |
PREDICTED: uncharacterized protein LOC105643970 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001205_240 |
0.0882258877 |
- |
- |
PREDICTED: uncharacterized protein LOC105645528 [Jatropha curcas] |
| 19 |
Hb_000484_130 |
0.0882673415 |
- |
- |
splicing factor, putative [Ricinus communis] |
| 20 |
Hb_007811_050 |
0.0888159996 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase NAK [Jatropha curcas] |