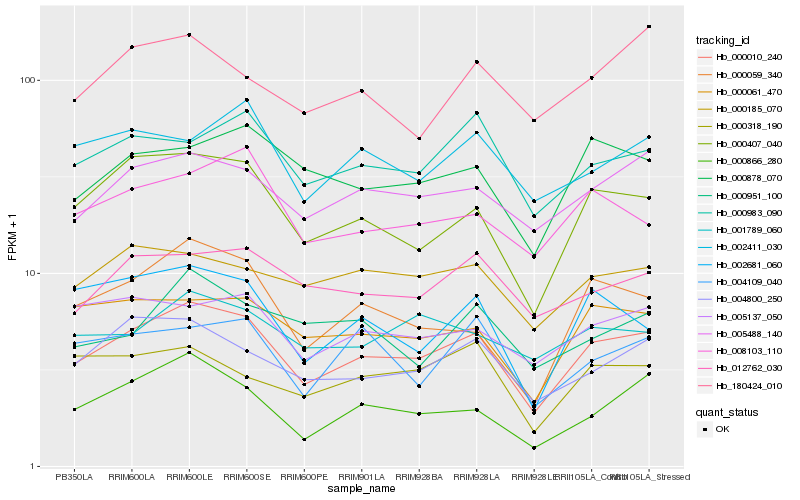
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000010_240 |
0.0 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 2 |
Hb_000407_040 |
0.0844646835 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_005488_140 |
0.0951781215 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 4 |
Hb_004800_250 |
0.0988625204 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A [Jatropha curcas] |
| 5 |
Hb_000059_340 |
0.104816043 |
transcription factor |
TF Family: SOH1 |
uncharacterized protein LOC100527879 [Glycine max] |
| 6 |
Hb_000061_470 |
0.1062198876 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: DNA annealing helicase and endonuclease ZRANB3 [Jatropha curcas] |
| 7 |
Hb_000983_090 |
0.108436344 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 8 |
Hb_012762_030 |
0.1087346001 |
- |
- |
PREDICTED: uncharacterized protein LOC105634062 [Jatropha curcas] |
| 9 |
Hb_000878_070 |
0.1095773731 |
- |
- |
hypothetical protein POPTR_0016s06670g [Populus trichocarpa] |
| 10 |
Hb_008103_110 |
0.1121142281 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_000866_280 |
0.1121795936 |
- |
- |
PREDICTED: seven transmembrane domain-containing tyrosine-protein kinase 1-like [Jatropha curcas] |
| 12 |
Hb_000185_070 |
0.1124338997 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005137_050 |
0.1127749579 |
- |
- |
PREDICTED: nuclear pore complex protein NUP58 [Jatropha curcas] |
| 14 |
Hb_002681_060 |
0.1174747227 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 15 |
Hb_002411_030 |
0.1186469141 |
transcription factor |
TF Family: NF-YC |
PREDICTED: dr1-associated corepressor homolog [Jatropha curcas] |
| 16 |
Hb_001789_060 |
0.1190300428 |
- |
- |
calmodulin-binding heat-shock protein, putative [Ricinus communis] |
| 17 |
Hb_004109_040 |
0.1196083291 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g53600, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000318_190 |
0.1211342134 |
- |
- |
hypothetical protein JCGZ_04889 [Jatropha curcas] |
| 19 |
Hb_000951_100 |
0.1231344906 |
- |
- |
PREDICTED: uncharacterized protein LOC103436565 isoform X1 [Malus domestica] |
| 20 |
Hb_180424_010 |
0.1240278725 |
- |
- |
Charged multivesicular body protein, putative [Ricinus communis] |