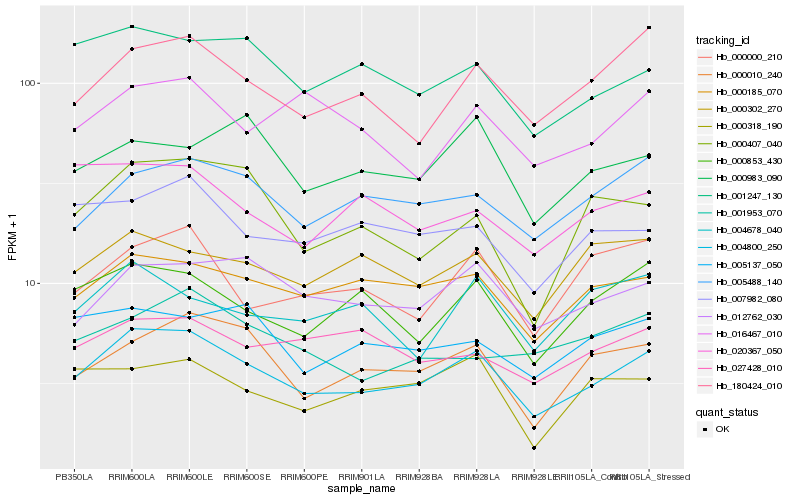
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004800_250 |
0.0 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A [Jatropha curcas] |
| 2 |
Hb_000185_070 |
0.0961724851 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_180424_010 |
0.0975364476 |
- |
- |
Charged multivesicular body protein, putative [Ricinus communis] |
| 4 |
Hb_000010_240 |
0.0988625204 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 5 |
Hb_012762_030 |
0.1006679684 |
- |
- |
PREDICTED: uncharacterized protein LOC105634062 [Jatropha curcas] |
| 6 |
Hb_005488_140 |
0.1017942885 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 7 |
Hb_000853_430 |
0.1020203472 |
- |
- |
PREDICTED: uncharacterized protein LOC105642597 [Jatropha curcas] |
| 8 |
Hb_001247_130 |
0.1068872638 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha [Jatropha curcas] |
| 9 |
Hb_000000_210 |
0.110119768 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_007982_080 |
0.112035869 |
- |
- |
PREDICTED: uncharacterized protein LOC105636531 [Jatropha curcas] |
| 11 |
Hb_005137_050 |
0.114361537 |
- |
- |
PREDICTED: nuclear pore complex protein NUP58 [Jatropha curcas] |
| 12 |
Hb_000407_040 |
0.1148854158 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_000302_270 |
0.1151744032 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_000318_190 |
0.1174765574 |
- |
- |
hypothetical protein JCGZ_04889 [Jatropha curcas] |
| 15 |
Hb_016467_010 |
0.1193990671 |
- |
- |
hypothetical protein POPTR_0004s18340g [Populus trichocarpa] |
| 16 |
Hb_001953_070 |
0.1200889433 |
- |
- |
PREDICTED: RNA pseudouridine synthase 7 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000983_090 |
0.1210757877 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 18 |
Hb_027428_010 |
0.1217112836 |
- |
- |
alpha-1,3-mannosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_004678_040 |
0.1217691929 |
- |
- |
PREDICTED: uncharacterized CRM domain-containing protein At3g25440, chloroplastic [Jatropha curcas] |
| 20 |
Hb_020367_050 |
0.1221006721 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl pyrophosphate synthase, putative [Ricinus communis] |