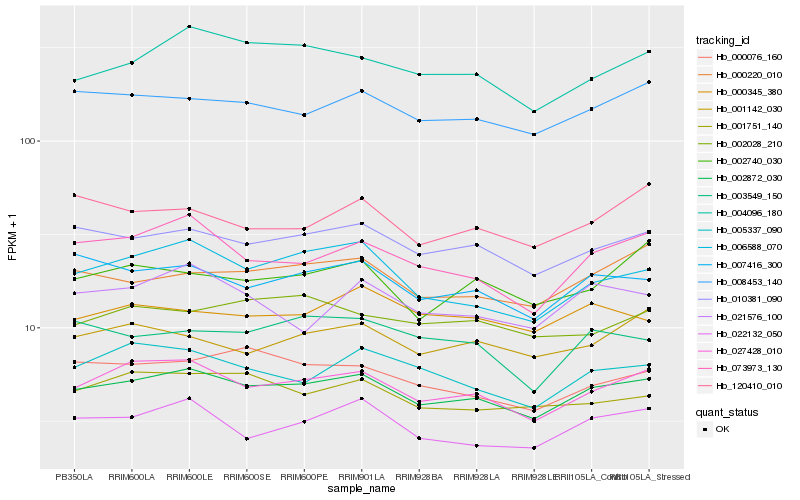
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002872_030 |
0.0 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 2 |
Hb_027428_010 |
0.0414312833 |
- |
- |
alpha-1,3-mannosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_010381_090 |
0.0441075059 |
- |
- |
PREDICTED: protein FLX-like 3 [Jatropha curcas] |
| 4 |
Hb_008453_140 |
0.0538880753 |
- |
- |
PREDICTED: SKP1-like protein 1B [Jatropha curcas] |
| 5 |
Hb_007416_300 |
0.0580998595 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 5b-like [Jatropha curcas] |
| 6 |
Hb_006588_070 |
0.0606870185 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 7 |
Hb_073973_130 |
0.0623536737 |
- |
- |
PREDICTED: prolyl-tRNA synthetase associated domain-containing protein 1 [Jatropha curcas] |
| 8 |
Hb_004096_180 |
0.0628046008 |
- |
- |
PREDICTED: 14-3-3-like protein A [Jatropha curcas] |
| 9 |
Hb_000220_010 |
0.0633785119 |
- |
- |
PREDICTED: uncharacterized protein LOC105630083 [Jatropha curcas] |
| 10 |
Hb_022132_050 |
0.0657582947 |
- |
- |
GTP binding protein, putative [Ricinus communis] |
| 11 |
Hb_001751_140 |
0.066548744 |
- |
- |
PREDICTED: uncharacterized protein LOC105648510 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005337_090 |
0.0669280199 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOUSLED-like [Jatropha curcas] |
| 13 |
Hb_000076_160 |
0.0705866323 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 14 |
Hb_001142_030 |
0.0706105103 |
- |
- |
PREDICTED: uncharacterized protein LOC105640023 [Jatropha curcas] |
| 15 |
Hb_000345_380 |
0.0717446188 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 7, peroxisomal [Jatropha curcas] |
| 16 |
Hb_021576_100 |
0.0739796205 |
- |
- |
Enoyl-CoA hydratase, mitochondrial precursor, putative [Ricinus communis] |
| 17 |
Hb_003549_150 |
0.074056075 |
- |
- |
clathrin coat adaptor ap3 medium chain, putative [Ricinus communis] |
| 18 |
Hb_002740_030 |
0.0741950842 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_120410_010 |
0.0746957405 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727-like [Solanum tuberosum] |
| 20 |
Hb_002028_210 |
0.0747506204 |
- |
- |
PREDICTED: uncharacterized protein LOC105638038 [Jatropha curcas] |