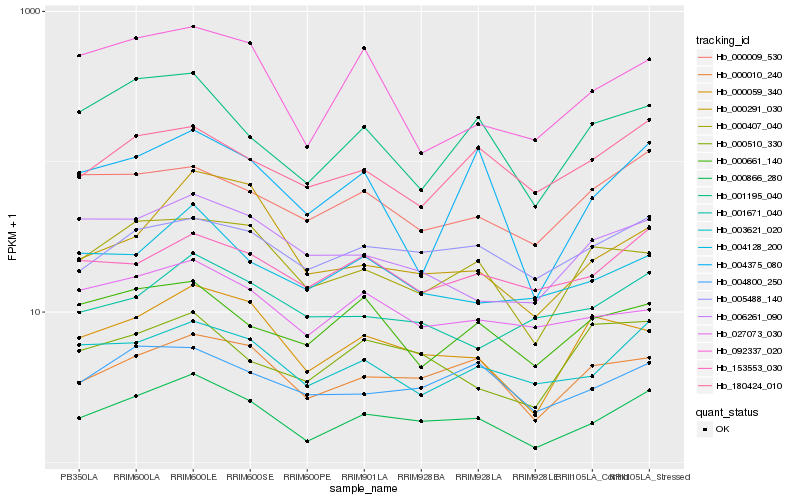
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000866_280 |
0.0 |
- |
- |
PREDICTED: seven transmembrane domain-containing tyrosine-protein kinase 1-like [Jatropha curcas] |
| 2 |
Hb_000010_240 |
0.1121795936 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 3 |
Hb_003621_020 |
0.123455204 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 4 |
Hb_000059_340 |
0.1265040599 |
transcription factor |
TF Family: SOH1 |
uncharacterized protein LOC100527879 [Glycine max] |
| 5 |
Hb_000407_040 |
0.1324847798 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_001195_040 |
0.1346183162 |
- |
- |
PREDICTED: proteasome subunit alpha type-1-B-like [Gossypium raimondii] |
| 7 |
Hb_004800_250 |
0.1349589524 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A [Jatropha curcas] |
| 8 |
Hb_005488_140 |
0.1421901791 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 9 |
Hb_004128_200 |
0.1424039306 |
- |
- |
PREDICTED: uncharacterized protein LOC105642137 [Jatropha curcas] |
| 10 |
Hb_000291_030 |
0.1459521032 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Eucalyptus grandis] |
| 11 |
Hb_004375_080 |
0.1476763424 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF073 [Jatropha curcas] |
| 12 |
Hb_180424_010 |
0.1480102447 |
- |
- |
Charged multivesicular body protein, putative [Ricinus communis] |
| 13 |
Hb_006261_090 |
0.1497286812 |
- |
- |
PREDICTED: exosome complex exonuclease RRP46 homolog [Jatropha curcas] |
| 14 |
Hb_000009_530 |
0.1501887309 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 15 |
Hb_153553_030 |
0.1503366626 |
- |
- |
PREDICTED: zinc finger protein GIS2 [Jatropha curcas] |
| 16 |
Hb_092337_020 |
0.1515106653 |
- |
- |
hypothetical protein B456_013G264900 [Gossypium raimondii] |
| 17 |
Hb_027073_030 |
0.1523565095 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 18 |
Hb_000661_140 |
0.1544618613 |
- |
- |
PREDICTED: uncharacterized protein LOC105648317 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001671_040 |
0.1547972114 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS31-like isoform X1 [Populus euphratica] |
| 20 |
Hb_000510_330 |
0.155913692 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |