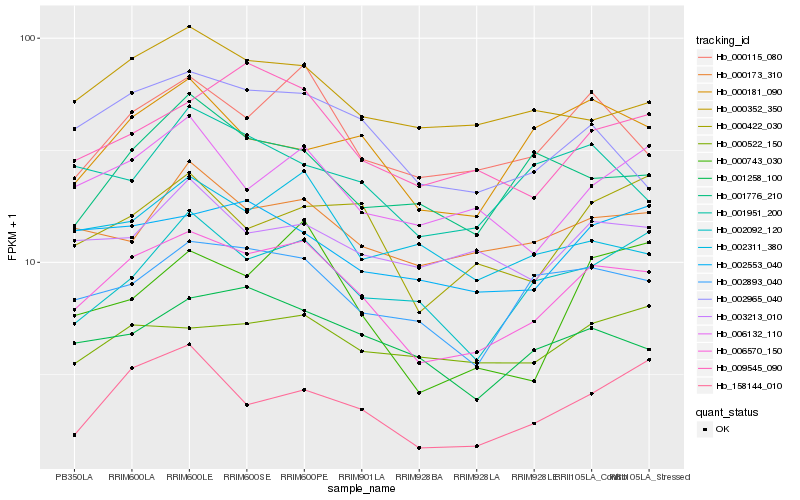
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006570_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635342 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000115_080 |
0.0993897299 |
- |
- |
PREDICTED: uncharacterized protein LOC105642032 [Jatropha curcas] |
| 3 |
Hb_002893_040 |
0.1023318767 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 6 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002965_040 |
0.1093153724 |
- |
- |
drought-responsive family protein [Populus trichocarpa] |
| 5 |
Hb_001258_100 |
0.1095815872 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_009545_090 |
0.1110466195 |
- |
- |
hypothetical protein JCGZ_02186 [Jatropha curcas] |
| 7 |
Hb_002092_120 |
0.1133023006 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000173_310 |
0.1140034923 |
- |
- |
PREDICTED: uncharacterized protein LOC105631893 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000352_350 |
0.1141306141 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000422_030 |
0.1144821194 |
- |
- |
PREDICTED: uncharacterized protein LOC105643943 [Jatropha curcas] |
| 11 |
Hb_006132_110 |
0.1166980646 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002553_040 |
0.1177068915 |
- |
- |
PREDICTED: transcription factor bHLH104 [Jatropha curcas] |
| 13 |
Hb_002311_380 |
0.1225774234 |
- |
- |
PREDICTED: induced during hyphae development protein 1 [Jatropha curcas] |
| 14 |
Hb_000522_150 |
0.1240380632 |
- |
- |
PREDICTED: uncharacterized protein LOC105646670 [Jatropha curcas] |
| 15 |
Hb_003213_010 |
0.1240891235 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 16 |
Hb_158144_010 |
0.1244630764 |
- |
- |
- |
| 17 |
Hb_000181_090 |
0.1251237531 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 18 |
Hb_000743_030 |
0.1267595034 |
- |
- |
NmrA-like negative transcriptional regulator family protein [Theobroma cacao] |
| 19 |
Hb_001776_210 |
0.1274335622 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 20 |
Hb_001951_200 |
0.12807493 |
- |
- |
PREDICTED: cold-regulated 413 inner membrane protein 1, chloroplastic-like [Jatropha curcas] |