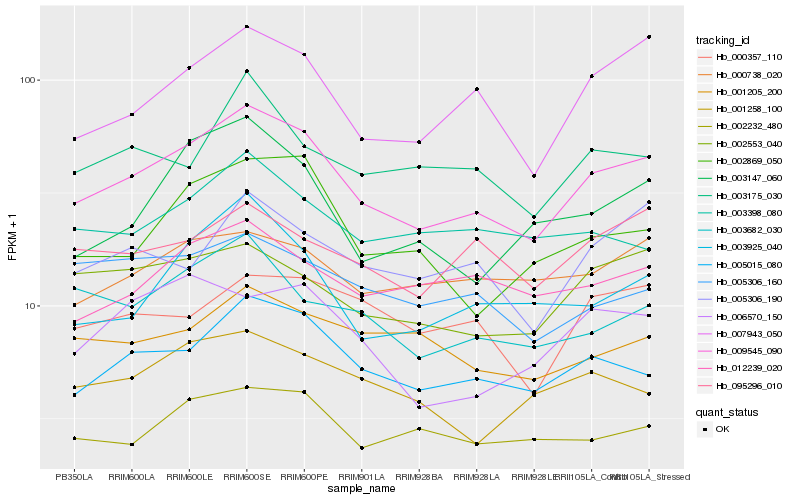
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009545_090 |
0.0 |
- |
- |
hypothetical protein JCGZ_02186 [Jatropha curcas] |
| 2 |
Hb_005015_080 |
0.0750341717 |
- |
- |
PREDICTED: shikimate kinase 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_007943_050 |
0.0885635877 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 4 |
Hb_002232_480 |
0.0982794045 |
- |
- |
PREDICTED: telomerase reverse transcriptase [Jatropha curcas] |
| 5 |
Hb_002869_050 |
0.0987405516 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 6 |
Hb_003925_040 |
0.0991569256 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger AN1 and C2H2 domain-containing stress-associated protein 16 [Jatropha curcas] |
| 7 |
Hb_002553_040 |
0.1002651777 |
- |
- |
PREDICTED: transcription factor bHLH104 [Jatropha curcas] |
| 8 |
Hb_003682_030 |
0.1024518764 |
- |
- |
hypothetical protein CICLE_v10020876mg [Citrus clementina] |
| 9 |
Hb_012239_020 |
0.103514769 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 10 |
Hb_003147_060 |
0.1047400261 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |
| 11 |
Hb_000738_020 |
0.1048626398 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_005306_190 |
0.1053376367 |
- |
- |
Mitochondrial import inner membrane translocase subunit tim23, putative [Ricinus communis] |
| 13 |
Hb_003175_030 |
0.106132851 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 14 |
Hb_001258_100 |
0.1062352827 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_001205_200 |
0.1087157711 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000357_110 |
0.1095529038 |
- |
- |
PREDICTED: uncharacterized protein LOC105643970 isoform X2 [Jatropha curcas] |
| 17 |
Hb_095296_010 |
0.109633775 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 18 |
Hb_005306_160 |
0.110616036 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1-like [Jatropha curcas] |
| 19 |
Hb_003398_080 |
0.110840858 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 20 |
Hb_006570_150 |
0.1110466195 |
- |
- |
PREDICTED: uncharacterized protein LOC105635342 isoform X1 [Jatropha curcas] |