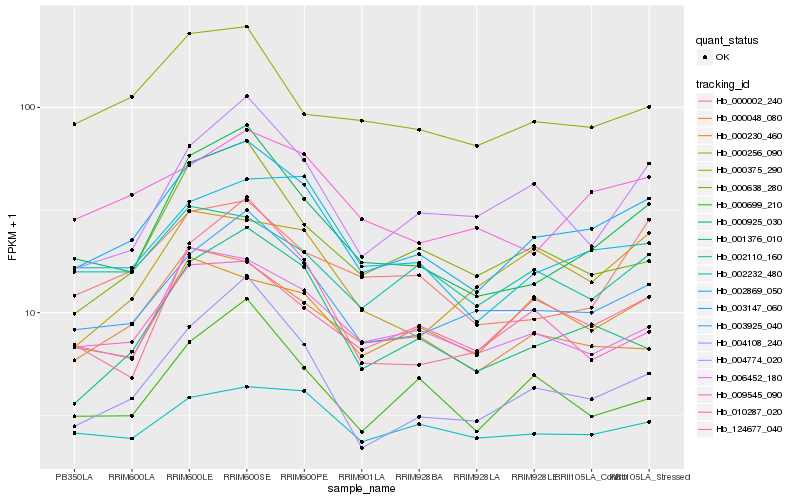
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003147_060 |
0.0 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |
| 2 |
Hb_003925_040 |
0.0772185661 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger AN1 and C2H2 domain-containing stress-associated protein 16 [Jatropha curcas] |
| 3 |
Hb_000925_030 |
0.0866491421 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 4 |
Hb_002869_050 |
0.096075521 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 5 |
Hb_006452_180 |
0.1024941647 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 6 |
Hb_004108_240 |
0.1029580206 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 7 |
Hb_009545_090 |
0.1047400261 |
- |
- |
hypothetical protein JCGZ_02186 [Jatropha curcas] |
| 8 |
Hb_001376_010 |
0.1089487831 |
- |
- |
diacylglycerol kinase, alpha, putative [Ricinus communis] |
| 9 |
Hb_000048_080 |
0.1092665909 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004774_020 |
0.1092975083 |
- |
- |
PREDICTED: PRA1 family protein B4-like [Camelina sativa] |
| 11 |
Hb_000002_240 |
0.1097323676 |
- |
- |
PREDICTED: transcription factor bHLH104 [Jatropha curcas] |
| 12 |
Hb_010287_020 |
0.1123694006 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 13 |
Hb_000375_290 |
0.1124835009 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_000230_460 |
0.1125662363 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |
| 15 |
Hb_002110_160 |
0.1142891587 |
- |
- |
PREDICTED: phosphoribosylaminoimidazole-succinocarboxamide synthase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_124677_040 |
0.1145461966 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 17 |
Hb_000699_210 |
0.1153626952 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 18 |
Hb_000638_280 |
0.115885497 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 19 |
Hb_002232_480 |
0.1174154076 |
- |
- |
PREDICTED: telomerase reverse transcriptase [Jatropha curcas] |
| 20 |
Hb_000256_090 |
0.1186192298 |
- |
- |
PREDICTED: uncharacterized protein LOC105637579 [Jatropha curcas] |