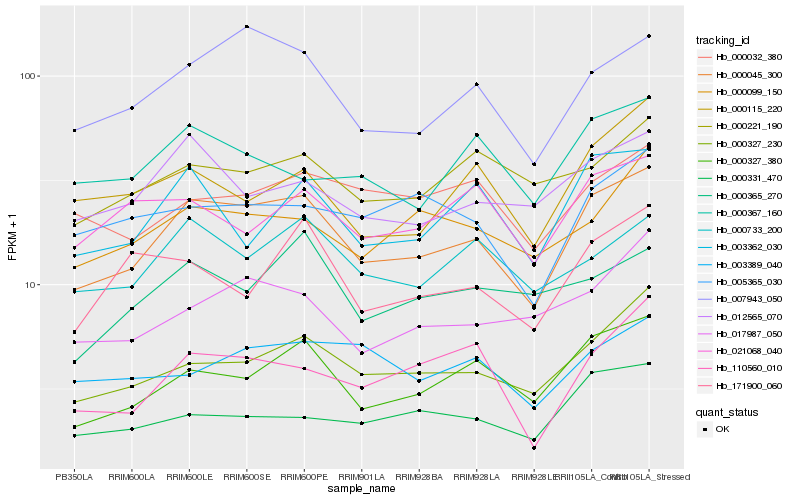
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000045_300 |
0.0 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier isoform X1 [Jatropha curcas] |
| 2 |
Hb_007943_050 |
0.0882005483 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 3 |
Hb_000327_380 |
0.092379381 |
- |
- |
hypothetical protein JCGZ_06621 [Jatropha curcas] |
| 4 |
Hb_000327_230 |
0.0960166321 |
- |
- |
PREDICTED: nitrilase-like protein 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000733_200 |
0.109492002 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 4 [Jatropha curcas] |
| 6 |
Hb_003362_030 |
0.1121093091 |
- |
- |
hypothetical protein POPTR_0017s08440g [Populus trichocarpa] |
| 7 |
Hb_012565_070 |
0.114883603 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 8 |
Hb_017987_050 |
0.1164319508 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 5, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_000099_150 |
0.1164339545 |
- |
- |
PREDICTED: calcium-dependent protein kinase 3 [Jatropha curcas] |
| 10 |
Hb_005365_030 |
0.1193789788 |
- |
- |
hypothetical protein POPTR_0002s24010g [Populus trichocarpa] |
| 11 |
Hb_110560_010 |
0.1202062086 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_000221_190 |
0.1216668547 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 13 |
Hb_000115_220 |
0.1224506527 |
- |
- |
hypothetical protein CISIN_1g0379742mg, partial [Citrus sinensis] |
| 14 |
Hb_000032_380 |
0.1224771471 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_171900_060 |
0.1225157974 |
- |
- |
PREDICTED: uncharacterized protein LOC105643368 [Jatropha curcas] |
| 16 |
Hb_000331_470 |
0.1225682764 |
- |
- |
PREDICTED: uncharacterized protein LOC105136689 [Populus euphratica] |
| 17 |
Hb_000367_160 |
0.1228084728 |
- |
- |
PREDICTED: SNAP25 homologous protein SNAP33 [Populus euphratica] |
| 18 |
Hb_021068_040 |
0.1229813658 |
- |
- |
PREDICTED: XIAP-associated factor 1 [Jatropha curcas] |
| 19 |
Hb_003389_040 |
0.1250002229 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 12 [Jatropha curcas] |
| 20 |
Hb_000365_270 |
0.1250384302 |
- |
- |
PREDICTED: uncharacterized protein LOC105649060 [Jatropha curcas] |