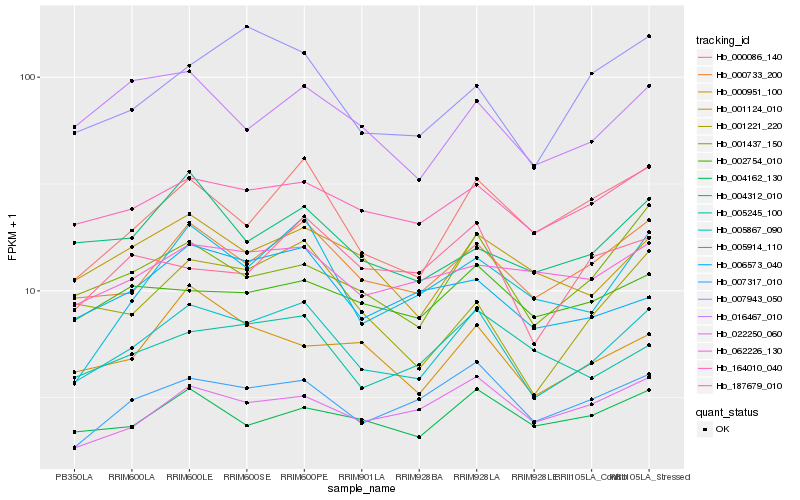
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005867_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 2 |
Hb_000733_200 |
0.077699164 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 4 [Jatropha curcas] |
| 3 |
Hb_007317_010 |
0.0921548132 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_164010_040 |
0.0963678502 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 5 |
Hb_000086_140 |
0.099838441 |
- |
- |
PREDICTED: probable arabinose 5-phosphate isomerase [Jatropha curcas] |
| 6 |
Hb_001124_010 |
0.1026924534 |
- |
- |
PREDICTED: serine/threonine-protein kinase WNK8 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001437_150 |
0.10397348 |
- |
- |
PREDICTED: SNF1-related protein kinase regulatory subunit beta-3-like [Jatropha curcas] |
| 8 |
Hb_006573_040 |
0.1055111332 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_022250_060 |
0.1072290539 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_001221_220 |
0.1080949359 |
- |
- |
PREDICTED: deoxycytidylate deaminase isoform X1 [Jatropha curcas] |
| 11 |
Hb_005914_110 |
0.1083456824 |
- |
- |
PREDICTED: uncharacterized protein LOC105632180 [Jatropha curcas] |
| 12 |
Hb_004162_130 |
0.1111277395 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005245_100 |
0.1114614062 |
- |
- |
PREDICTED: uncharacterized protein At4g08330, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_016467_010 |
0.1120564171 |
- |
- |
hypothetical protein POPTR_0004s18340g [Populus trichocarpa] |
| 15 |
Hb_000951_100 |
0.1122119399 |
- |
- |
PREDICTED: uncharacterized protein LOC103436565 isoform X1 [Malus domestica] |
| 16 |
Hb_007943_050 |
0.1132925309 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 17 |
Hb_002754_010 |
0.1140215466 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 18 |
Hb_062226_130 |
0.115714437 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 4 [Jatropha curcas] |
| 19 |
Hb_187679_010 |
0.1160672746 |
- |
- |
- |
| 20 |
Hb_004312_010 |
0.117543816 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X2 [Jatropha curcas] |