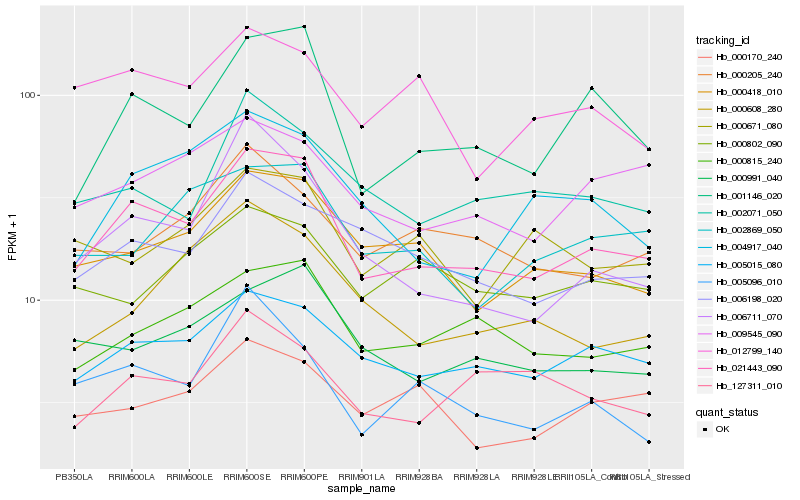
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021443_090 |
0.0 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 2 |
Hb_001146_020 |
0.0960769003 |
- |
- |
hypothetical protein CISIN_1g013079mg [Citrus sinensis] |
| 3 |
Hb_005015_080 |
0.0963405044 |
- |
- |
PREDICTED: shikimate kinase 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000815_240 |
0.1116729397 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000418_010 |
0.1140793467 |
- |
- |
PREDICTED: repressor of RNA polymerase III transcription MAF1 homolog [Jatropha curcas] |
| 6 |
Hb_000991_040 |
0.1203081164 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 7 |
Hb_006198_020 |
0.1209492743 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 8 |
Hb_009545_090 |
0.1241284554 |
- |
- |
hypothetical protein JCGZ_02186 [Jatropha curcas] |
| 9 |
Hb_002071_050 |
0.1250751496 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 5-like [Jatropha curcas] |
| 10 |
Hb_004917_040 |
0.1258608802 |
- |
- |
hypothetical protein POPTR_0009s03470g [Populus trichocarpa] |
| 11 |
Hb_006711_070 |
0.1277522159 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 12 |
Hb_002869_050 |
0.1293706881 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 13 |
Hb_127311_010 |
0.1348714215 |
- |
- |
hypothetical protein JCGZ_07497 [Jatropha curcas] |
| 14 |
Hb_000205_240 |
0.1348767137 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_000608_280 |
0.1360037496 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 16 |
Hb_000170_240 |
0.136396337 |
- |
- |
PREDICTED: auxilin-related protein 1 [Jatropha curcas] |
| 17 |
Hb_012799_140 |
0.1374708507 |
- |
- |
PREDICTED: membrane steroid-binding protein 2-like [Jatropha curcas] |
| 18 |
Hb_005096_010 |
0.1388188478 |
- |
- |
PREDICTED: uncharacterized protein LOC105641403 [Jatropha curcas] |
| 19 |
Hb_000671_080 |
0.1409888694 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 20 |
Hb_000802_090 |
0.1411507818 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |