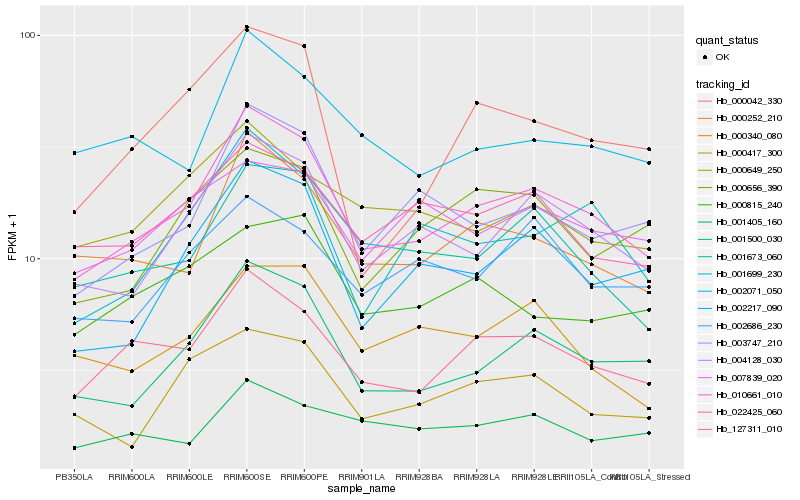
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010661_010 |
0.0 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 2 |
Hb_004128_030 |
0.0793487596 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 8-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_001500_030 |
0.0805322742 |
- |
- |
PREDICTED: uncharacterized protein At2g33490 isoform X2 [Jatropha curcas] |
| 4 |
Hb_127311_010 |
0.0805488522 |
- |
- |
hypothetical protein JCGZ_07497 [Jatropha curcas] |
| 5 |
Hb_000252_210 |
0.0899119938 |
- |
- |
PREDICTED: uncharacterized protein LOC105644655 [Jatropha curcas] |
| 6 |
Hb_003747_210 |
0.0966872054 |
- |
- |
PREDICTED: uncharacterized protein LOC105630240 [Jatropha curcas] |
| 7 |
Hb_001673_060 |
0.1018581606 |
- |
- |
PREDICTED: uncharacterized protein LOC105635789 [Jatropha curcas] |
| 8 |
Hb_002217_090 |
0.1018612552 |
- |
- |
PREDICTED: probable protein phosphatase 2C 13 [Jatropha curcas] |
| 9 |
Hb_001699_230 |
0.1057820889 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |
| 10 |
Hb_002071_050 |
0.1095120682 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 5-like [Jatropha curcas] |
| 11 |
Hb_022425_060 |
0.1133251774 |
- |
- |
PREDICTED: protein phosphatase 2C and cyclic nucleotide-binding/kinase domain-containing protein isoform X1 [Jatropha curcas] |
| 12 |
Hb_000340_080 |
0.1175195295 |
- |
- |
PREDICTED: isoamylase 2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000417_300 |
0.1185838477 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7 [Jatropha curcas] |
| 14 |
Hb_000649_250 |
0.1190277918 |
transcription factor |
TF Family: C2H2 |
PREDICTED: UBX domain-containing protein 6 [Jatropha curcas] |
| 15 |
Hb_000656_390 |
0.1205308295 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000042_330 |
0.1207142949 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002686_230 |
0.122794051 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 18 |
Hb_000815_240 |
0.1228505651 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_007839_020 |
0.1255093145 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001405_160 |
0.1257473445 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g09680 [Jatropha curcas] |