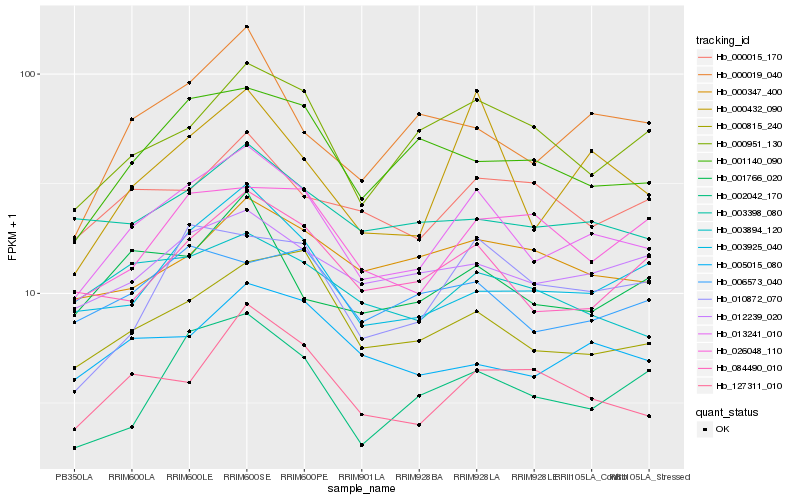
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013241_010 |
0.0 |
- |
- |
PREDICTED: small G protein signaling modulator 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000432_090 |
0.1043746104 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 3 |
Hb_084490_010 |
0.1067253337 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000815_240 |
0.1097346155 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_003925_040 |
0.1099072422 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger AN1 and C2H2 domain-containing stress-associated protein 16 [Jatropha curcas] |
| 6 |
Hb_127311_010 |
0.1104239813 |
- |
- |
hypothetical protein JCGZ_07497 [Jatropha curcas] |
| 7 |
Hb_012239_020 |
0.1132930777 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 8 |
Hb_000951_130 |
0.1156204362 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 9 |
Hb_010872_070 |
0.1157585008 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001140_090 |
0.1177515278 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002042_170 |
0.1192299877 |
- |
- |
RING/U-box superfamily protein [Theobroma cacao] |
| 12 |
Hb_005015_080 |
0.1197679446 |
- |
- |
PREDICTED: shikimate kinase 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003894_120 |
0.1205484892 |
- |
- |
PREDICTED: uncharacterized protein LOC105643110 [Jatropha curcas] |
| 14 |
Hb_001766_020 |
0.1229772378 |
- |
- |
PREDICTED: probable galacturonosyltransferase 14 [Populus euphratica] |
| 15 |
Hb_003398_080 |
0.123994655 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 16 |
Hb_000019_040 |
0.1240588178 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 3, mitochondrial [Jatropha curcas] |
| 17 |
Hb_006573_040 |
0.1270990536 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_000015_170 |
0.1273692675 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000347_400 |
0.1285295991 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_026048_110 |
0.1286775329 |
- |
- |
PREDICTED: uncharacterized protein LOC105645733 [Jatropha curcas] |