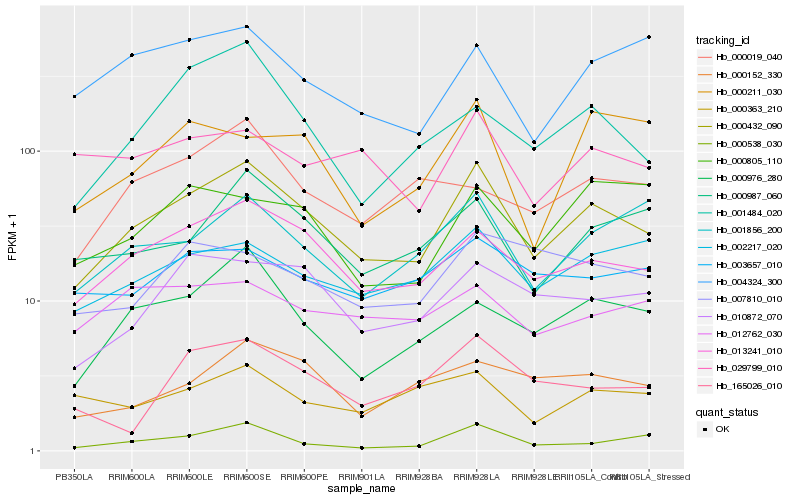
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000432_090 |
0.0 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 2 |
Hb_013241_010 |
0.1043746104 |
- |
- |
PREDICTED: small G protein signaling modulator 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_010872_070 |
0.144678136 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000976_280 |
0.1446989163 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13-like [Jatropha curcas] |
| 5 |
Hb_000987_060 |
0.1456888634 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 6 |
Hb_002217_020 |
0.1487216417 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001856_200 |
0.1507561638 |
- |
- |
Esterase PIR7B, putative [Ricinus communis] |
| 8 |
Hb_000538_030 |
0.1527523203 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 9 |
Hb_000152_330 |
0.158657101 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 10 |
Hb_001484_020 |
0.1607284698 |
- |
- |
Phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_003657_010 |
0.164204359 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_165026_010 |
0.1644854934 |
transcription factor |
TF Family: NAC |
hypothetical protein JCGZ_14166 [Jatropha curcas] |
| 13 |
Hb_004324_300 |
0.165999415 |
- |
- |
hypothetical protein PRUPE_ppa012628mg [Prunus persica] |
| 14 |
Hb_029799_010 |
0.166951567 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 15 |
Hb_007810_010 |
0.1713688135 |
- |
- |
PREDICTED: uncharacterized protein LOC105647655 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000805_110 |
0.1715246476 |
- |
- |
PREDICTED: uncharacterized protein C24B11.05 [Jatropha curcas] |
| 17 |
Hb_000363_210 |
0.1720127115 |
- |
- |
PREDICTED: putative MO25-like protein At5g47540 [Jatropha curcas] |
| 18 |
Hb_012762_030 |
0.1724139828 |
- |
- |
PREDICTED: uncharacterized protein LOC105634062 [Jatropha curcas] |
| 19 |
Hb_000019_040 |
0.1726013021 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 3, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000211_030 |
0.174131788 |
- |
- |
sugar transporter, putative [Ricinus communis] |