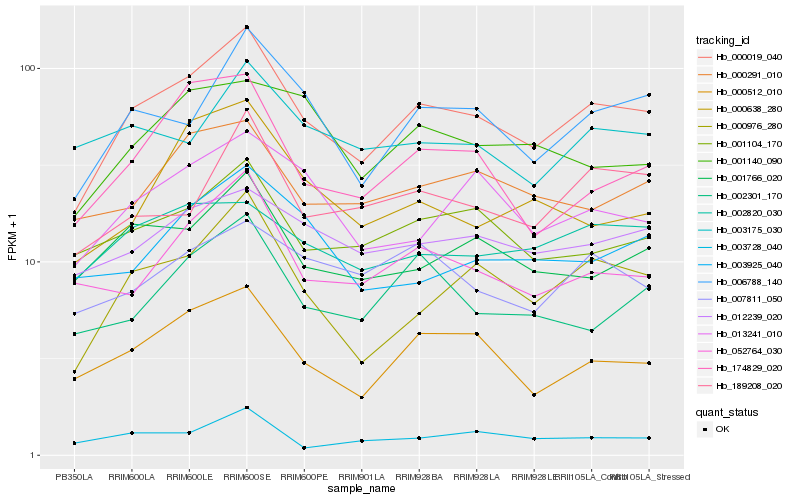
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000019_040 |
0.0 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 3, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000976_280 |
0.0983087021 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13-like [Jatropha curcas] |
| 3 |
Hb_006788_140 |
0.0994175805 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |
| 4 |
Hb_000512_010 |
0.1033293905 |
- |
- |
PREDICTED: uncharacterized protein LOC105650728 [Jatropha curcas] |
| 5 |
Hb_001766_020 |
0.1143852614 |
- |
- |
PREDICTED: probable galacturonosyltransferase 14 [Populus euphratica] |
| 6 |
Hb_052764_030 |
0.1148219463 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 7 |
Hb_001104_170 |
0.115881018 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2c [Jatropha curcas] |
| 8 |
Hb_174829_020 |
0.1205563935 |
transcription factor |
TF Family: C2C2-Dof |
cyclic dof factor 2 [Jatropha curcas] |
| 9 |
Hb_012239_020 |
0.1216081825 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 10 |
Hb_000291_010 |
0.1221405007 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 11 |
Hb_189208_020 |
0.1232251538 |
- |
- |
LRX2, putative [Ricinus communis] |
| 12 |
Hb_013241_010 |
0.1240588178 |
- |
- |
PREDICTED: small G protein signaling modulator 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002301_170 |
0.1246801374 |
- |
- |
MAC/Perforin domain-containing protein [Theobroma cacao] |
| 14 |
Hb_002820_030 |
0.1303544853 |
- |
- |
PREDICTED: uncharacterized protein LOC105638922 [Jatropha curcas] |
| 15 |
Hb_007811_050 |
0.1316087416 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase NAK [Jatropha curcas] |
| 16 |
Hb_003925_040 |
0.1325482482 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger AN1 and C2H2 domain-containing stress-associated protein 16 [Jatropha curcas] |
| 17 |
Hb_003728_040 |
0.1329197424 |
- |
- |
PREDICTED: uncharacterized protein LOC101492305 [Cicer arietinum] |
| 18 |
Hb_001140_090 |
0.1338857016 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003175_030 |
0.1354784268 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 20 |
Hb_000638_280 |
0.1365964127 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |