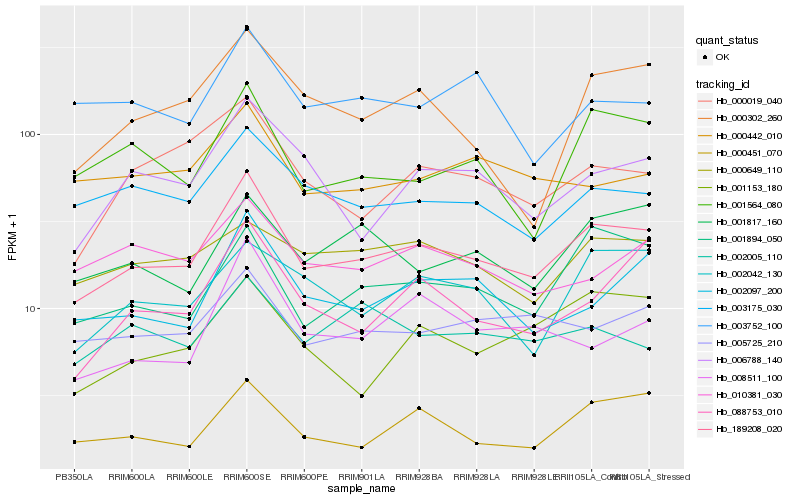
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_189208_020 |
0.0 |
- |
- |
LRX2, putative [Ricinus communis] |
| 2 |
Hb_002097_200 |
0.1092271998 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 3 |
Hb_001894_050 |
0.1097039038 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 2 [Jatropha curcas] |
| 4 |
Hb_003175_030 |
0.1104211839 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 5 |
Hb_005725_210 |
0.1145395506 |
- |
- |
PREDICTED: RNA-binding protein 1 [Jatropha curcas] |
| 6 |
Hb_006788_140 |
0.1190978685 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |
| 7 |
Hb_001564_080 |
0.1206599857 |
transcription factor |
TF Family: NF-YB |
PREDICTED: protein Dr1 homolog isoform X2 [Jatropha curcas] |
| 8 |
Hb_000451_070 |
0.1212764125 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 9 |
Hb_001817_160 |
0.1228490031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000019_040 |
0.1232251538 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 3, mitochondrial [Jatropha curcas] |
| 11 |
Hb_002042_130 |
0.1244264792 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 12 |
Hb_010381_030 |
0.1257757811 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 13 |
Hb_088753_010 |
0.1270852284 |
- |
- |
PREDICTED: copper amine oxidase 1-like [Vitis vinifera] |
| 14 |
Hb_000442_010 |
0.127169988 |
- |
- |
hypothetical protein POPTR_0010s19060g [Populus trichocarpa] |
| 15 |
Hb_000302_260 |
0.127714211 |
- |
- |
hypothetical protein [Bruguiera gymnorhiza] |
| 16 |
Hb_000649_110 |
0.1277160282 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 17 |
Hb_001153_180 |
0.131162235 |
- |
- |
PREDICTED: SPX domain-containing membrane protein At4g22990-like [Jatropha curcas] |
| 18 |
Hb_008511_100 |
0.1319765012 |
- |
- |
hypothetical protein POPTR_0009s11860g [Populus trichocarpa] |
| 19 |
Hb_003752_100 |
0.1319816563 |
- |
- |
PREDICTED: FRIGIDA-like protein 3 [Jatropha curcas] |
| 20 |
Hb_002005_110 |
0.1324095164 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X5 [Jatropha curcas] |