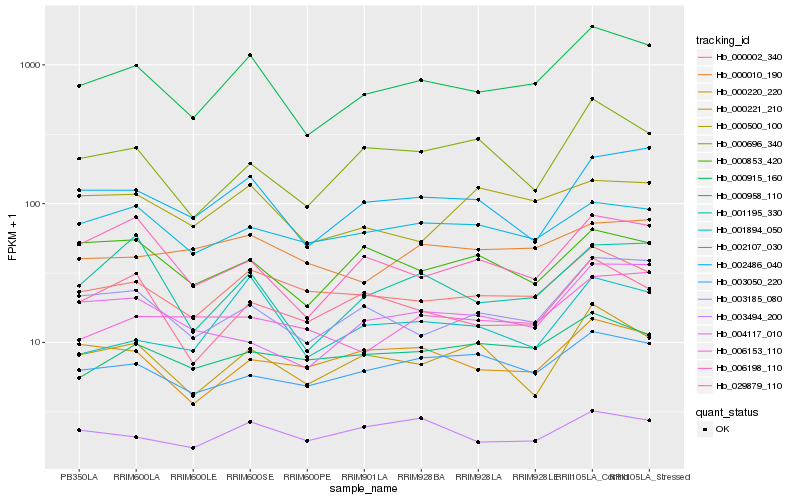
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000915_160 |
0.0 |
- |
- |
PREDICTED: histone H1 [Jatropha curcas] |
| 2 |
Hb_003185_080 |
0.1015030523 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 3 |
Hb_002486_040 |
0.1022480504 |
- |
- |
- |
| 4 |
Hb_000002_340 |
0.1057584645 |
- |
- |
PREDICTED: peroxisomal bifunctional enzyme-like [Jatropha curcas] |
| 5 |
Hb_001195_330 |
0.1151155222 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000221_210 |
0.1181456355 |
- |
- |
Kinase superfamily protein isoform 2 [Theobroma cacao] |
| 7 |
Hb_003050_220 |
0.1191878496 |
- |
- |
hypothetical protein JCGZ_04601 [Jatropha curcas] |
| 8 |
Hb_001894_050 |
0.1212157082 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 2 [Jatropha curcas] |
| 9 |
Hb_006198_110 |
0.1218088419 |
- |
- |
PREDICTED: uncharacterized protein At4g13200, chloroplastic [Jatropha curcas] |
| 10 |
Hb_029879_110 |
0.1251453481 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g61870, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000220_220 |
0.1254200976 |
- |
- |
PREDICTED: uncharacterized protein LOC105636907 [Jatropha curcas] |
| 12 |
Hb_000696_340 |
0.1259540122 |
- |
- |
PREDICTED: uncharacterized protein LOC105647255 [Jatropha curcas] |
| 13 |
Hb_000958_110 |
0.1268234849 |
- |
- |
PREDICTED: uncharacterized protein LOC105628719 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002107_030 |
0.1281246831 |
- |
- |
PREDICTED: COPII coat assembly protein SEC16 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003494_200 |
0.1285744693 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15-like [Jatropha curcas] |
| 16 |
Hb_004117_010 |
0.1324109267 |
- |
- |
60S ribosomal protein L10, mitochondrial, putative [Ricinus communis] |
| 17 |
Hb_000500_100 |
0.1324618421 |
- |
- |
peptide methionine sulfoxide reductase A1-like [Jatropha curcas] |
| 18 |
Hb_006153_110 |
0.1331119364 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 19 |
Hb_000853_420 |
0.1343342165 |
- |
- |
PREDICTED: phosphoribosylformylglycinamidine cyclo-ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 20 |
Hb_000010_190 |
0.1359256616 |
- |
- |
PREDICTED: protein-L-isoaspartate O-methyltransferase 1 isoform X2 [Jatropha curcas] |