| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
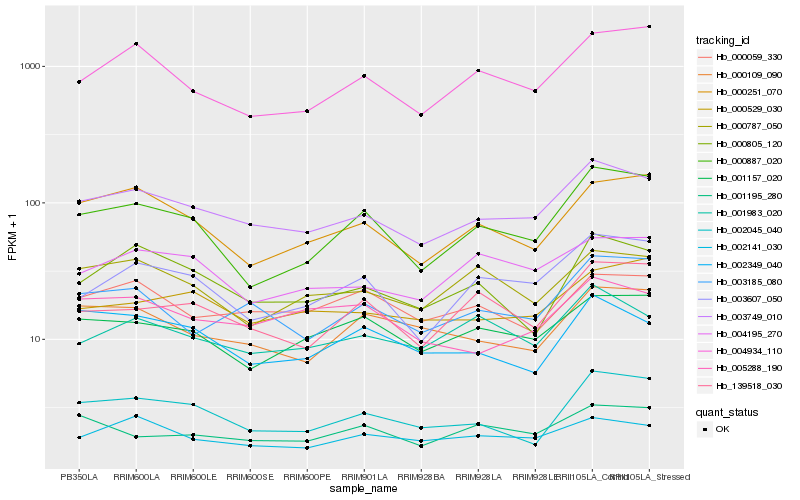
Hb_003749_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003607_050 |
0.0901173281 |
- |
- |
PREDICTED: uncharacterized protein LOC105644428 [Jatropha curcas] |
| 3 |
Hb_001983_020 |
0.090550461 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Jatropha curcas] |
| 4 |
Hb_003185_080 |
0.0914886825 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 5 |
Hb_001157_020 |
0.0916645209 |
- |
- |
PREDICTED: uncharacterized protein LOC105649844 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002141_030 |
0.0930487752 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004195_270 |
0.0944179924 |
- |
- |
Ubiquitin-conjugating enzyme 16 [Theobroma cacao] |
| 8 |
Hb_000251_070 |
0.0946941654 |
- |
- |
hypothetical protein L484_010641 [Morus notabilis] |
| 9 |
Hb_000805_120 |
0.0956340045 |
- |
- |
ubiquitin fusion degradaton protein, putative [Ricinus communis] |
| 10 |
Hb_004934_110 |
0.097993517 |
transcription factor |
TF Family: HMG |
hypothetical protein B456_002G115100 [Gossypium raimondii] |
| 11 |
Hb_000059_330 |
0.0981168304 |
- |
- |
PREDICTED: polyadenylate-binding protein 1 [Jatropha curcas] |
| 12 |
Hb_000787_050 |
0.0985097306 |
- |
- |
PREDICTED: electron transfer flavoprotein subunit alpha, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_000529_030 |
0.0985767786 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_005288_190 |
0.0985935153 |
- |
- |
PREDICTED: exportin-4 [Jatropha curcas] |
| 15 |
Hb_000109_090 |
0.0989635736 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 16 |
Hb_002045_040 |
0.1017002185 |
- |
- |
PREDICTED: protein OSB2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_002349_040 |
0.1025604943 |
- |
- |
PREDICTED: RNA pseudouridine synthase 5 [Jatropha curcas] |
| 18 |
Hb_000887_020 |
0.1033007611 |
- |
- |
PREDICTED: uncharacterized protein LOC105633527 [Jatropha curcas] |
| 19 |
Hb_139518_030 |
0.1041660434 |
- |
- |
protein kinase atn1, putative [Ricinus communis] |
| 20 |
Hb_001195_280 |
0.1043140833 |
- |
- |
PREDICTED: uncharacterized protein LOC103928873 isoform X1 [Pyrus x bretschneideri] |