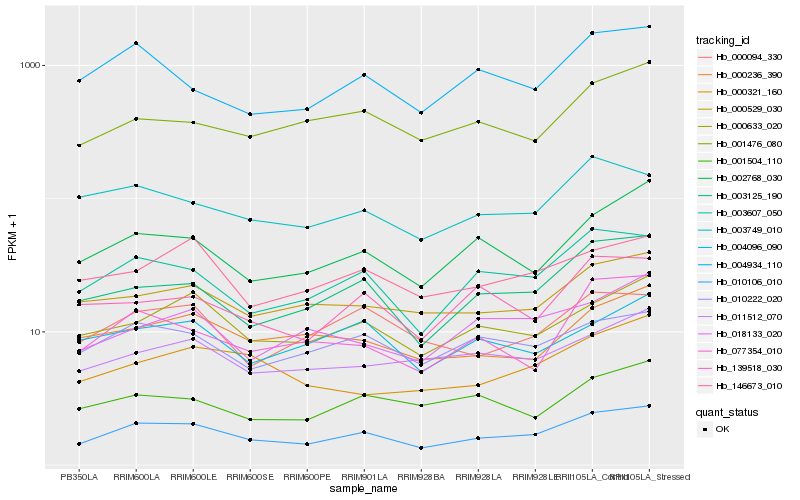
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010106_010 |
0.0 |
- |
- |
PREDICTED: cucumisin-like [Populus euphratica] |
| 2 |
Hb_003607_050 |
0.0803891249 |
- |
- |
PREDICTED: uncharacterized protein LOC105644428 [Jatropha curcas] |
| 3 |
Hb_003125_190 |
0.0879342565 |
- |
- |
hypothetical protein POPTR_0015s10960g [Populus trichocarpa] |
| 4 |
Hb_000633_020 |
0.0897566582 |
- |
- |
PREDICTED: uncharacterized protein LOC105630002 [Jatropha curcas] |
| 5 |
Hb_000529_030 |
0.096558236 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_010222_020 |
0.101310669 |
- |
- |
PREDICTED: uncharacterized protein LOC105647642 [Jatropha curcas] |
| 7 |
Hb_002768_030 |
0.1014991921 |
- |
- |
gaba(A) receptor-associated protein, putative [Ricinus communis] |
| 8 |
Hb_011512_070 |
0.1043623242 |
transcription factor |
TF Family: PHD |
Inhibitor of growth protein, putative [Ricinus communis] |
| 9 |
Hb_000236_390 |
0.1063852063 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH1 [Jatropha curcas] |
| 10 |
Hb_001476_080 |
0.1064260125 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 5A-like [Jatropha curcas] |
| 11 |
Hb_146673_010 |
0.1067639941 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 12 |
Hb_004934_110 |
0.1070414284 |
transcription factor |
TF Family: HMG |
hypothetical protein B456_002G115100 [Gossypium raimondii] |
| 13 |
Hb_000094_330 |
0.1100614928 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 4 [Jatropha curcas] |
| 14 |
Hb_003749_010 |
0.1101559314 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001504_110 |
0.110982469 |
- |
- |
PREDICTED: uncharacterized protein LOC105630336 [Jatropha curcas] |
| 16 |
Hb_004096_090 |
0.1121618627 |
- |
- |
Plastid-specific 30S ribosomal protein 3, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_139518_030 |
0.1122184714 |
- |
- |
protein kinase atn1, putative [Ricinus communis] |
| 18 |
Hb_000321_160 |
0.1125913299 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |
| 19 |
Hb_018133_020 |
0.1132420448 |
- |
- |
transporter, putative [Ricinus communis] |
| 20 |
Hb_077354_010 |
0.1134939398 |
- |
- |
PREDICTED: uncharacterized protein LOC105637567 [Jatropha curcas] |