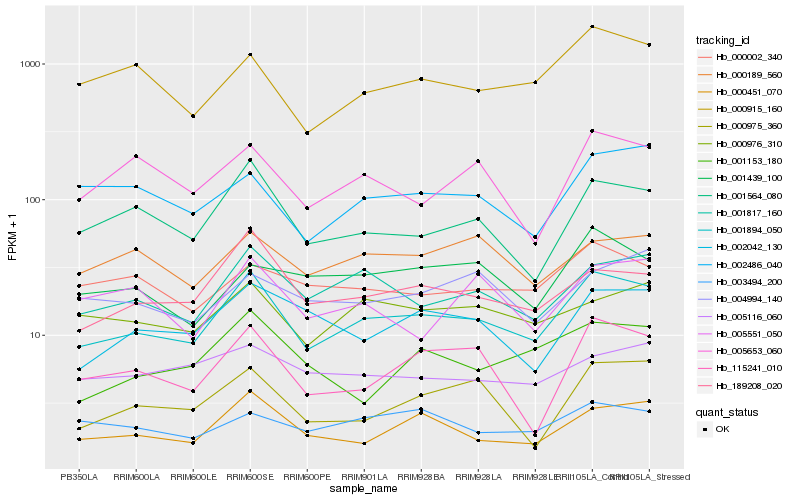
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001894_050 |
0.0 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 2 [Jatropha curcas] |
| 2 |
Hb_001817_160 |
0.1035731952 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001564_080 |
0.1086336705 |
transcription factor |
TF Family: NF-YB |
PREDICTED: protein Dr1 homolog isoform X2 [Jatropha curcas] |
| 4 |
Hb_189208_020 |
0.1097039038 |
- |
- |
LRX2, putative [Ricinus communis] |
| 5 |
Hb_000451_070 |
0.1169188752 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 6 |
Hb_000915_160 |
0.1212157082 |
- |
- |
PREDICTED: histone H1 [Jatropha curcas] |
| 7 |
Hb_002042_130 |
0.1268173081 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 8 |
Hb_115241_010 |
0.1274114946 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC [Jatropha curcas] |
| 9 |
Hb_000976_310 |
0.1294183155 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005653_060 |
0.1332079684 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 3 [Jatropha curcas] |
| 11 |
Hb_002486_040 |
0.1339146799 |
- |
- |
- |
| 12 |
Hb_000975_360 |
0.1363950228 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001439_100 |
0.1384822463 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 14 |
Hb_000002_340 |
0.1387700671 |
- |
- |
PREDICTED: peroxisomal bifunctional enzyme-like [Jatropha curcas] |
| 15 |
Hb_001153_180 |
0.1388003593 |
- |
- |
PREDICTED: SPX domain-containing membrane protein At4g22990-like [Jatropha curcas] |
| 16 |
Hb_000189_560 |
0.1391676504 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1 [Jatropha curcas] |
| 17 |
Hb_003494_200 |
0.1413457128 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15-like [Jatropha curcas] |
| 18 |
Hb_005551_050 |
0.1436833953 |
- |
- |
PREDICTED: nudix hydrolase 27, chloroplastic [Jatropha curcas] |
| 19 |
Hb_005116_060 |
0.1447746592 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004994_140 |
0.1456236753 |
- |
- |
PREDICTED: digalactosyldiacylglycerol synthase 2, chloroplastic [Jatropha curcas] |