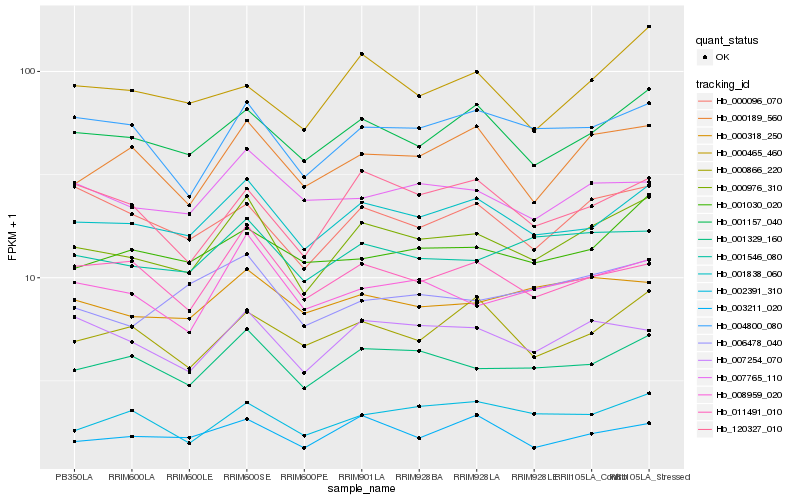
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000976_310 |
0.0 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001838_060 |
0.0608785936 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38)-like isoform X3 [Jatropha curcas] |
| 3 |
Hb_001329_160 |
0.0648512338 |
- |
- |
PREDICTED: uncharacterized protein LOC105642679 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001157_040 |
0.0743480344 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 5 |
Hb_008959_020 |
0.0806567824 |
- |
- |
PREDICTED: nitric oxide synthase-interacting protein [Pyrus x bretschneideri] |
| 6 |
Hb_001546_080 |
0.081650602 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12b isoform X1 [Jatropha curcas] |
| 7 |
Hb_000189_560 |
0.0850625772 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1 [Jatropha curcas] |
| 8 |
Hb_004800_080 |
0.0857717381 |
- |
- |
PREDICTED: acidic leucine-rich nuclear phosphoprotein 32-related protein [Jatropha curcas] |
| 9 |
Hb_007254_070 |
0.0866834807 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_011491_010 |
0.0901439236 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 11 |
Hb_006478_040 |
0.0915658318 |
- |
- |
PREDICTED: methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000318_250 |
0.0921022676 |
- |
- |
PREDICTED: uncharacterized protein LOC105634251 [Jatropha curcas] |
| 13 |
Hb_002391_310 |
0.092709884 |
- |
- |
hypothetical protein PRUPE_ppa021315mg [Prunus persica] |
| 14 |
Hb_001030_020 |
0.0929904409 |
- |
- |
Poly(A) polymerase alpha, putative [Ricinus communis] |
| 15 |
Hb_000096_070 |
0.0936697439 |
- |
- |
PREDICTED: ruvB-like 2 [Jatropha curcas] |
| 16 |
Hb_003211_020 |
0.093867011 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g08310, mitochondrial [Jatropha curcas] |
| 17 |
Hb_120327_010 |
0.0945802746 |
- |
- |
PREDICTED: uncharacterized protein LOC105650959 [Jatropha curcas] |
| 18 |
Hb_000866_220 |
0.0952924126 |
- |
- |
PREDICTED: uncharacterized protein LOC105642989 [Jatropha curcas] |
| 19 |
Hb_000465_460 |
0.0955125655 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 25 [Jatropha curcas] |
| 20 |
Hb_007765_110 |
0.0958337582 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 40-like [Jatropha curcas] |