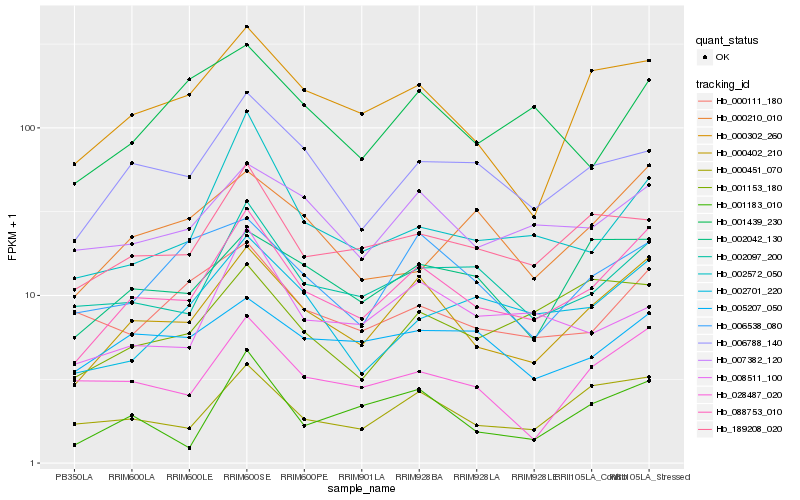
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_088753_010 |
0.0 |
- |
- |
PREDICTED: copper amine oxidase 1-like [Vitis vinifera] |
| 2 |
Hb_000402_210 |
0.0624658735 |
- |
- |
hypothetical protein EUGRSUZ_B032971, partial [Eucalyptus grandis] |
| 3 |
Hb_002097_200 |
0.1161764011 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 4 |
Hb_000451_070 |
0.1205431982 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 5 |
Hb_189208_020 |
0.1270852284 |
- |
- |
LRX2, putative [Ricinus communis] |
| 6 |
Hb_006788_140 |
0.1285537932 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |
| 7 |
Hb_000302_260 |
0.1307074656 |
- |
- |
hypothetical protein [Bruguiera gymnorhiza] |
| 8 |
Hb_007382_120 |
0.1328975735 |
- |
- |
PREDICTED: delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial [Jatropha curcas] |
| 9 |
Hb_002701_220 |
0.1370507744 |
- |
- |
PREDICTED: solute carrier family 25 member 44 [Jatropha curcas] |
| 10 |
Hb_002572_050 |
0.1379514287 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_028487_020 |
0.1424736068 |
- |
- |
hypothetical protein POPTR_0010s11880g [Populus trichocarpa] |
| 12 |
Hb_002042_130 |
0.1446462974 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 13 |
Hb_001183_010 |
0.1506579183 |
- |
- |
- |
| 14 |
Hb_001153_180 |
0.1516904524 |
- |
- |
PREDICTED: SPX domain-containing membrane protein At4g22990-like [Jatropha curcas] |
| 15 |
Hb_000210_010 |
0.1532559642 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 16 |
Hb_000111_180 |
0.1543989038 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 17 |
Hb_005207_050 |
0.1547005204 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 39 [Jatropha curcas] |
| 18 |
Hb_006538_080 |
0.1564511642 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 19 |
Hb_008511_100 |
0.1574076055 |
- |
- |
hypothetical protein POPTR_0009s11860g [Populus trichocarpa] |
| 20 |
Hb_001439_230 |
0.1580437487 |
- |
- |
PREDICTED: cystathionine gamma-synthase 1, chloroplastic [Jatropha curcas] |