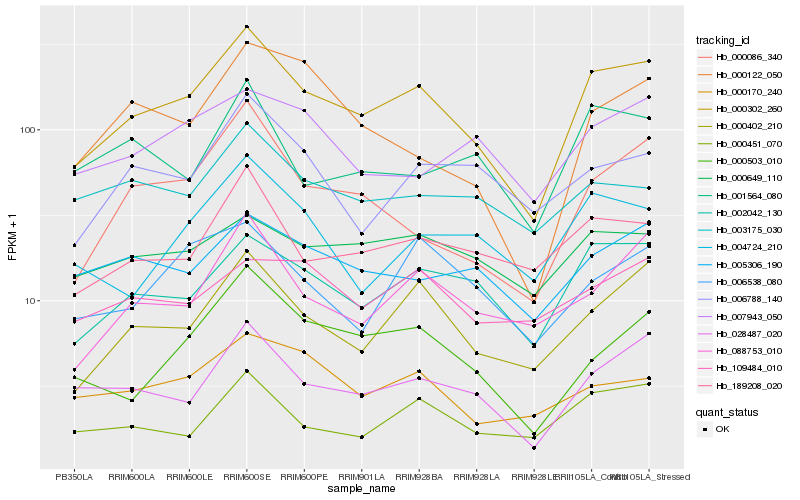
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000302_260 |
0.0 |
- |
- |
hypothetical protein [Bruguiera gymnorhiza] |
| 2 |
Hb_002042_130 |
0.1089783759 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 3 |
Hb_000402_210 |
0.1100185646 |
- |
- |
hypothetical protein EUGRSUZ_B032971, partial [Eucalyptus grandis] |
| 4 |
Hb_028487_020 |
0.1190924765 |
- |
- |
hypothetical protein POPTR_0010s11880g [Populus trichocarpa] |
| 5 |
Hb_189208_020 |
0.127714211 |
- |
- |
LRX2, putative [Ricinus communis] |
| 6 |
Hb_000086_340 |
0.1292788046 |
- |
- |
PREDICTED: uncharacterized protein LOC100253191 [Vitis vinifera] |
| 7 |
Hb_088753_010 |
0.1307074656 |
- |
- |
PREDICTED: copper amine oxidase 1-like [Vitis vinifera] |
| 8 |
Hb_000503_010 |
0.1331279519 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b [Jatropha curcas] |
| 9 |
Hb_000451_070 |
0.1337400346 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 10 |
Hb_000170_240 |
0.1423643253 |
- |
- |
PREDICTED: auxilin-related protein 1 [Jatropha curcas] |
| 11 |
Hb_004724_210 |
0.142665746 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005306_190 |
0.1428587388 |
- |
- |
Mitochondrial import inner membrane translocase subunit tim23, putative [Ricinus communis] |
| 13 |
Hb_006538_080 |
0.1441435098 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 14 |
Hb_000649_110 |
0.146210766 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 15 |
Hb_006788_140 |
0.1466939351 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |
| 16 |
Hb_007943_050 |
0.1504149766 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 17 |
Hb_001564_080 |
0.1506430729 |
transcription factor |
TF Family: NF-YB |
PREDICTED: protein Dr1 homolog isoform X2 [Jatropha curcas] |
| 18 |
Hb_003175_030 |
0.1534187308 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 19 |
Hb_000122_050 |
0.154528923 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_109484_010 |
0.1550439784 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |