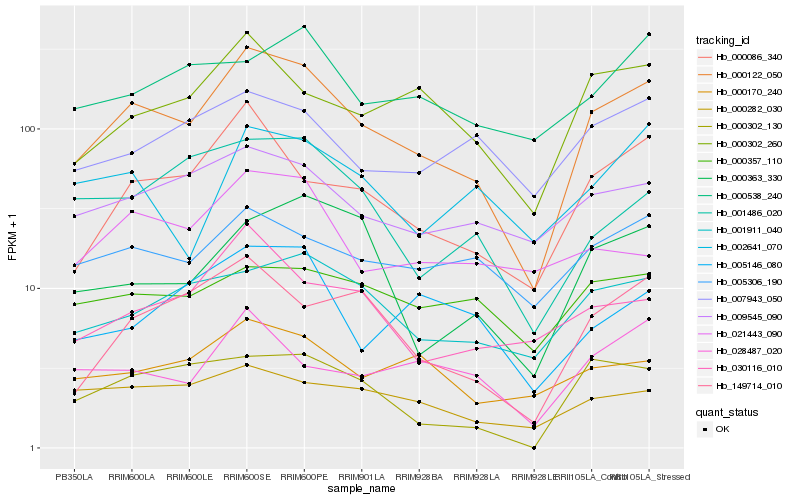
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000122_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000086_340 |
0.1388386989 |
- |
- |
PREDICTED: uncharacterized protein LOC100253191 [Vitis vinifera] |
| 3 |
Hb_001911_040 |
0.1535717722 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 4 |
Hb_000302_260 |
0.154528923 |
- |
- |
hypothetical protein [Bruguiera gymnorhiza] |
| 5 |
Hb_149714_010 |
0.1545997277 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like [Jatropha curcas] |
| 6 |
Hb_000302_130 |
0.1588628161 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |
| 7 |
Hb_009545_090 |
0.1605980523 |
- |
- |
hypothetical protein JCGZ_02186 [Jatropha curcas] |
| 8 |
Hb_000282_030 |
0.1674313671 |
- |
- |
K(+) efflux antiporter 4 [Glycine soja] |
| 9 |
Hb_000170_240 |
0.1674933803 |
- |
- |
PREDICTED: auxilin-related protein 1 [Jatropha curcas] |
| 10 |
Hb_005306_190 |
0.1679948183 |
- |
- |
Mitochondrial import inner membrane translocase subunit tim23, putative [Ricinus communis] |
| 11 |
Hb_030116_010 |
0.1698772598 |
- |
- |
F-box protein, atfbl3, putative [Ricinus communis] |
| 12 |
Hb_001486_020 |
0.1732528984 |
- |
- |
hypothetical protein JCGZ_02719 [Jatropha curcas] |
| 13 |
Hb_000363_330 |
0.1742748578 |
- |
- |
- |
| 14 |
Hb_007943_050 |
0.1769909586 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 15 |
Hb_028487_020 |
0.1774214942 |
- |
- |
hypothetical protein POPTR_0010s11880g [Populus trichocarpa] |
| 16 |
Hb_021443_090 |
0.1777421021 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 17 |
Hb_005146_080 |
0.177986706 |
- |
- |
PREDICTED: uncharacterized protein LOC105636704 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000538_240 |
0.1800197193 |
- |
- |
PREDICTED: uncharacterized protein LOC105643073 [Jatropha curcas] |
| 19 |
Hb_002641_070 |
0.1816472867 |
- |
- |
PREDICTED: transcription factor bHLH147-like [Jatropha curcas] |
| 20 |
Hb_000357_110 |
0.1822682269 |
- |
- |
PREDICTED: uncharacterized protein LOC105643970 isoform X2 [Jatropha curcas] |