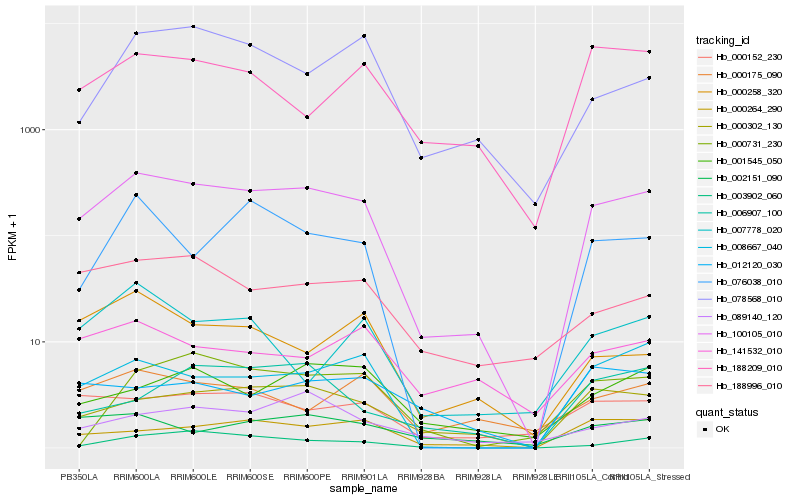
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_100105_010 |
0.0 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 2 |
Hb_000302_130 |
0.1548585384 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |
| 3 |
Hb_008667_040 |
0.1767857322 |
- |
- |
glycerol-3-phosphate transporter family protein [Populus trichocarpa] |
| 4 |
Hb_000264_290 |
0.1835711326 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB2 [Jatropha curcas] |
| 5 |
Hb_000152_230 |
0.1882477378 |
- |
- |
PREDICTED: uncharacterized protein LOC105648566 [Jatropha curcas] |
| 6 |
Hb_188996_010 |
0.1920966385 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003902_060 |
0.1945126104 |
- |
- |
PREDICTED: seed biotin-containing protein SBP65 [Jatropha curcas] |
| 8 |
Hb_076038_010 |
0.1967799307 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001545_050 |
0.2028852507 |
- |
- |
PREDICTED: ras-related protein Rab7 [Nicotiana sylvestris] |
| 10 |
Hb_089140_120 |
0.2062706578 |
- |
- |
PREDICTED: MOB kinase activator-like 1 [Jatropha curcas] |
| 11 |
Hb_002151_090 |
0.2068143784 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 1-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_078568_010 |
0.2096828281 |
- |
- |
hypothetical protein PRUPE_ppa013245mg [Prunus persica] |
| 13 |
Hb_000175_090 |
0.2105808418 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000258_320 |
0.2112494987 |
- |
- |
- |
| 15 |
Hb_188209_010 |
0.2127031034 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 7B, mitochondrial-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_007778_020 |
0.2174503716 |
- |
- |
- |
| 17 |
Hb_141532_010 |
0.2176472025 |
- |
- |
PREDICTED: protein SMG9-like [Jatropha curcas] |
| 18 |
Hb_000731_230 |
0.2183885417 |
- |
- |
PREDICTED: abnormal spindle-like microcephaly-associated protein homolog [Jatropha curcas] |
| 19 |
Hb_006907_100 |
0.2204128356 |
- |
- |
PREDICTED: tropinone reductase-like 3 [Jatropha curcas] |
| 20 |
Hb_012120_030 |
0.221173398 |
- |
- |
- |