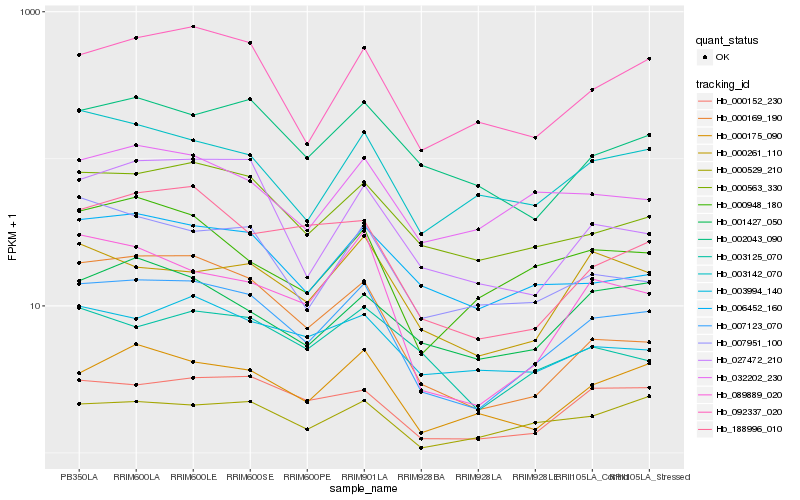
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007123_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_000152_230 |
0.1058735838 |
- |
- |
PREDICTED: uncharacterized protein LOC105648566 [Jatropha curcas] |
| 3 |
Hb_092337_020 |
0.1148540051 |
- |
- |
hypothetical protein B456_013G264900 [Gossypium raimondii] |
| 4 |
Hb_000175_090 |
0.116413924 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000563_330 |
0.1172975217 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_089889_020 |
0.1197626066 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At2g36080-like [Jatropha curcas] |
| 7 |
Hb_000529_210 |
0.128344605 |
- |
- |
PREDICTED: uncharacterized protein At4g38062 [Jatropha curcas] |
| 8 |
Hb_006452_160 |
0.1289899513 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 9 |
Hb_188996_010 |
0.1296084868 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003142_070 |
0.1314296074 |
- |
- |
hypothetical protein F383_14657 [Gossypium arboreum] |
| 11 |
Hb_007951_100 |
0.1329959989 |
- |
- |
PREDICTED: peptide deformylase 1A, chloroplastic/mitochondrial [Jatropha curcas] |
| 12 |
Hb_002043_090 |
0.1334531153 |
- |
- |
hypothetical protein JCGZ_24811 [Jatropha curcas] |
| 13 |
Hb_000169_190 |
0.1335623395 |
desease resistance |
Gene Name: AAA_17 |
PREDICTED: protein SMG9-like [Jatropha curcas] |
| 14 |
Hb_027472_210 |
0.1335644019 |
- |
- |
PREDICTED: uncharacterized protein LOC105646181 [Jatropha curcas] |
| 15 |
Hb_000948_180 |
0.1340764817 |
- |
- |
PREDICTED: uncharacterized protein LOC105629501 [Jatropha curcas] |
| 16 |
Hb_000261_110 |
0.1344160566 |
- |
- |
PREDICTED: probable methionine--tRNA ligase [Jatropha curcas] |
| 17 |
Hb_003994_140 |
0.1356041401 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_032202_230 |
0.1391410585 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 19 |
Hb_003125_070 |
0.1396789264 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio1-like [Jatropha curcas] |
| 20 |
Hb_001427_050 |
0.140674124 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |