| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
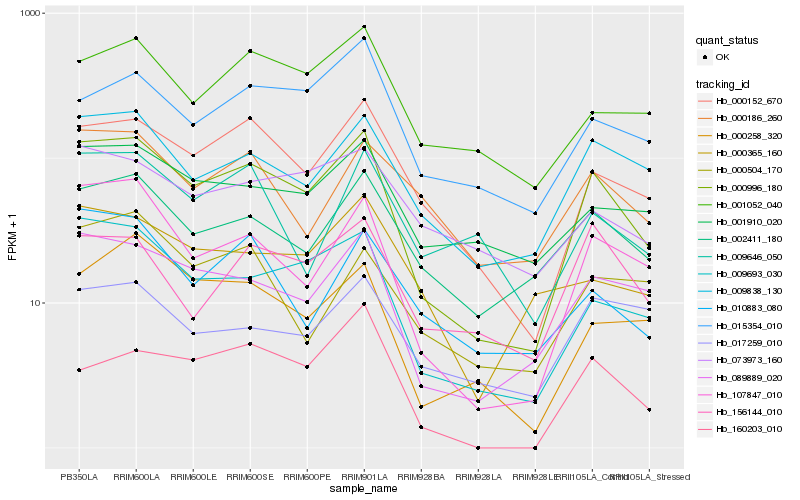
Hb_000996_180 |
0.0 |
- |
- |
PREDICTED: tetraspanin-10 [Jatropha curcas] |
| 2 |
Hb_107847_010 |
0.1190189837 |
- |
- |
3-isopropylmalate dehydratase, putative [Ricinus communis] |
| 3 |
Hb_089889_020 |
0.1209046428 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At2g36080-like [Jatropha curcas] |
| 4 |
Hb_009693_030 |
0.1209082813 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_000152_670 |
0.1221392875 |
- |
- |
hypothetical protein VITISV_037603 [Vitis vinifera] |
| 6 |
Hb_009838_130 |
0.1378393451 |
- |
- |
hypothetical protein CISIN_1g010403mg [Citrus sinensis] |
| 7 |
Hb_002411_180 |
0.147764764 |
- |
- |
PREDICTED: THO complex subunit 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000258_320 |
0.1538784885 |
- |
- |
- |
| 9 |
Hb_001052_040 |
0.161075791 |
- |
- |
Ferredoxin-3, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_001910_020 |
0.1634653588 |
- |
- |
PREDICTED: ubiquitin-associated domain-containing protein 2 [Jatropha curcas] |
| 11 |
Hb_015354_010 |
0.1650843628 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 12 |
Hb_156144_010 |
0.166352047 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9 [Jatropha curcas] |
| 13 |
Hb_160203_010 |
0.1709100081 |
- |
- |
- |
| 14 |
Hb_000365_160 |
0.1716292645 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 [Jatropha curcas] |
| 15 |
Hb_010883_080 |
0.1725709718 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHR1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000186_260 |
0.1736211602 |
transcription factor |
TF Family: PHD |
PREDICTED: protein polybromo-1-like [Jatropha curcas] |
| 17 |
Hb_073973_160 |
0.1758290574 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 18 |
Hb_009646_050 |
0.1779428286 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_017259_010 |
0.1781212463 |
- |
- |
- |
| 20 |
Hb_000504_170 |
0.179314831 |
- |
- |
hypothetical protein B456_006G033900 [Gossypium raimondii] |