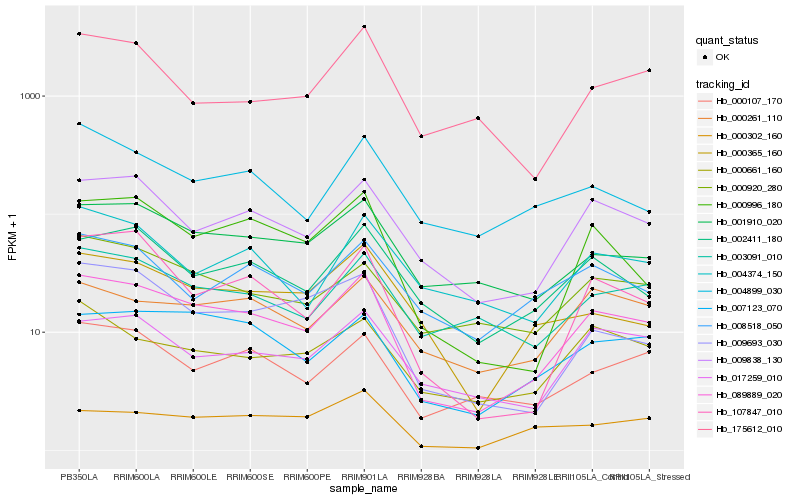
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_089889_020 |
0.0 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At2g36080-like [Jatropha curcas] |
| 2 |
Hb_009838_130 |
0.1151554669 |
- |
- |
hypothetical protein CISIN_1g010403mg [Citrus sinensis] |
| 3 |
Hb_007123_070 |
0.1197626066 |
- |
- |
- |
| 4 |
Hb_000996_180 |
0.1209046428 |
- |
- |
PREDICTED: tetraspanin-10 [Jatropha curcas] |
| 5 |
Hb_001910_020 |
0.1285555259 |
- |
- |
PREDICTED: ubiquitin-associated domain-containing protein 2 [Jatropha curcas] |
| 6 |
Hb_107847_010 |
0.132533075 |
- |
- |
3-isopropylmalate dehydratase, putative [Ricinus communis] |
| 7 |
Hb_017259_010 |
0.1340243742 |
- |
- |
- |
| 8 |
Hb_002411_180 |
0.1344007847 |
- |
- |
PREDICTED: THO complex subunit 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000107_170 |
0.1353696669 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC25 homolog [Jatropha curcas] |
| 10 |
Hb_009693_030 |
0.1357906387 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_000661_160 |
0.1389814104 |
- |
- |
hexokinase [Manihot esculenta] |
| 12 |
Hb_003091_010 |
0.1399658699 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 13 |
Hb_000261_110 |
0.1399733388 |
- |
- |
PREDICTED: probable methionine--tRNA ligase [Jatropha curcas] |
| 14 |
Hb_000365_160 |
0.1412496028 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 [Jatropha curcas] |
| 15 |
Hb_000920_280 |
0.1423494698 |
- |
- |
Protein SENSITIVITY TO RED LIGHT REDUCED, putative [Ricinus communis] |
| 16 |
Hb_000302_160 |
0.1512008725 |
- |
- |
PREDICTED: uncharacterized protein LOC105648403 [Jatropha curcas] |
| 17 |
Hb_004899_030 |
0.153651889 |
- |
- |
PREDICTED: 20 kDa chaperonin, chloroplastic [Jatropha curcas] |
| 18 |
Hb_008518_050 |
0.1563313789 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 19 |
Hb_004374_150 |
0.156645395 |
- |
- |
PREDICTED: uncharacterized protein LOC105650290 isoform X1 [Jatropha curcas] |
| 20 |
Hb_175612_010 |
0.1575321671 |
transcription factor |
TF Family: MBF1 |
orf [Ricinus communis] |