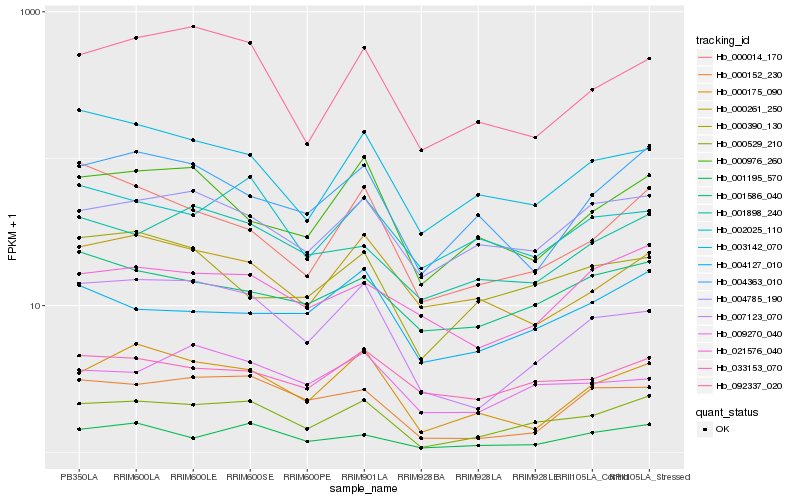
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000529_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At4g38062 [Jatropha curcas] |
| 2 |
Hb_001195_570 |
0.1257834087 |
- |
- |
PREDICTED: peroxisomal membrane protein 11A [Jatropha curcas] |
| 3 |
Hb_004785_190 |
0.1270783179 |
- |
- |
PHD finger family protein [Populus trichocarpa] |
| 4 |
Hb_007123_070 |
0.128344605 |
- |
- |
- |
| 5 |
Hb_000175_090 |
0.1321554484 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_092337_020 |
0.1337268532 |
- |
- |
hypothetical protein B456_013G264900 [Gossypium raimondii] |
| 7 |
Hb_000152_230 |
0.1369163766 |
- |
- |
PREDICTED: uncharacterized protein LOC105648566 [Jatropha curcas] |
| 8 |
Hb_001898_240 |
0.1375067263 |
- |
- |
PREDICTED: uncharacterized protein LOC105779688 [Gossypium raimondii] |
| 9 |
Hb_003142_070 |
0.1388334354 |
- |
- |
hypothetical protein F383_14657 [Gossypium arboreum] |
| 10 |
Hb_004363_010 |
0.1407278816 |
- |
- |
hypothetical protein CISIN_1g0405721mg, partial [Citrus sinensis] |
| 11 |
Hb_001586_040 |
0.1413291705 |
- |
- |
hypothetical protein JCGZ_17187 [Jatropha curcas] |
| 12 |
Hb_000976_260 |
0.1424637173 |
- |
- |
- |
| 13 |
Hb_033153_070 |
0.1425768741 |
- |
- |
PREDICTED: uncharacterized protein LOC105649606 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004127_010 |
0.1438756391 |
- |
- |
PREDICTED: probable RNA 3'-terminal phosphate cyclase-like protein [Malus domestica] |
| 15 |
Hb_000390_130 |
0.1443470645 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B isoform X2 [Jatropha curcas] |
| 16 |
Hb_021576_040 |
0.1446914707 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_009270_040 |
0.1454487867 |
- |
- |
- |
| 18 |
Hb_002025_110 |
0.1459312163 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: SWI/SNF complex component SNF12 homolog isoform X1 [Jatropha curcas] |
| 19 |
Hb_000261_250 |
0.1481162637 |
- |
- |
elongation factor 1-alpha [Lepidium sativum] |
| 20 |
Hb_000014_170 |
0.1501318987 |
- |
- |
hypothetical protein CISIN_1g0379742mg, partial [Citrus sinensis] |