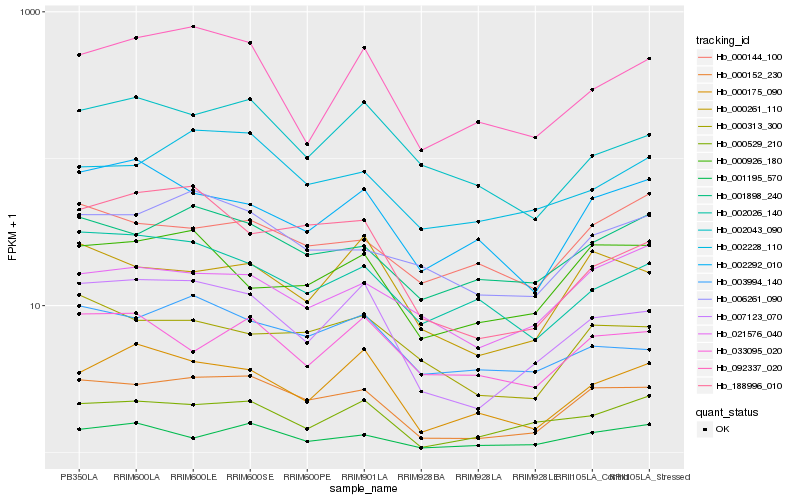
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_230 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648566 [Jatropha curcas] |
| 2 |
Hb_007123_070 |
0.1058735838 |
- |
- |
- |
| 3 |
Hb_001898_240 |
0.1277991271 |
- |
- |
PREDICTED: uncharacterized protein LOC105779688 [Gossypium raimondii] |
| 4 |
Hb_006261_090 |
0.1295329648 |
- |
- |
PREDICTED: exosome complex exonuclease RRP46 homolog [Jatropha curcas] |
| 5 |
Hb_000313_300 |
0.133419964 |
- |
- |
PREDICTED: F-box protein SKIP19-like [Jatropha curcas] |
| 6 |
Hb_000261_110 |
0.1335102981 |
- |
- |
PREDICTED: probable methionine--tRNA ligase [Jatropha curcas] |
| 7 |
Hb_000529_210 |
0.1369163766 |
- |
- |
PREDICTED: uncharacterized protein At4g38062 [Jatropha curcas] |
| 8 |
Hb_002228_110 |
0.1392823644 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |
| 9 |
Hb_000926_180 |
0.1406069345 |
- |
- |
proline synthetase associated protein, putative [Ricinus communis] |
| 10 |
Hb_001195_570 |
0.1434932619 |
- |
- |
PREDICTED: peroxisomal membrane protein 11A [Jatropha curcas] |
| 11 |
Hb_000175_090 |
0.1457891158 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003994_140 |
0.148013337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_021576_040 |
0.1496711977 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002292_010 |
0.1502167206 |
- |
- |
hypothetical protein CICLE_v10029628mg [Citrus clementina] |
| 15 |
Hb_092337_020 |
0.1507891725 |
- |
- |
hypothetical protein B456_013G264900 [Gossypium raimondii] |
| 16 |
Hb_188996_010 |
0.1509259043 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002043_090 |
0.1525954097 |
- |
- |
hypothetical protein JCGZ_24811 [Jatropha curcas] |
| 18 |
Hb_002026_140 |
0.1542939497 |
- |
- |
hypothetical protein POPTR_0006s13340g [Populus trichocarpa] |
| 19 |
Hb_000144_100 |
0.1545275789 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 2, mitochondrial [Jatropha curcas] |
| 20 |
Hb_033095_020 |
0.1557808578 |
- |
- |
conserved hypothetical protein [Ricinus communis] |