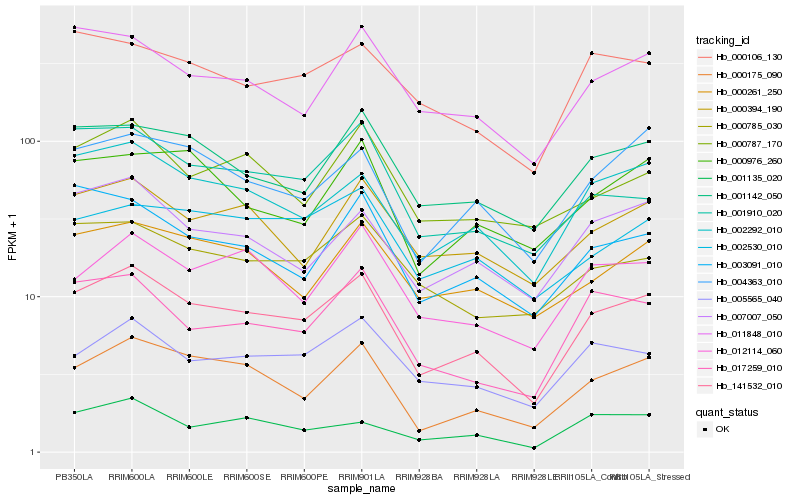
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_141532_010 |
0.0 |
- |
- |
PREDICTED: protein SMG9-like [Jatropha curcas] |
| 2 |
Hb_000175_090 |
0.0812482047 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002292_010 |
0.0820623324 |
- |
- |
hypothetical protein CICLE_v10029628mg [Citrus clementina] |
| 4 |
Hb_001142_050 |
0.0978723898 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |
| 5 |
Hb_004363_010 |
0.1012312157 |
- |
- |
hypothetical protein CISIN_1g0405721mg, partial [Citrus sinensis] |
| 6 |
Hb_005565_040 |
0.1018321623 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_007007_050 |
0.1022963207 |
- |
- |
PREDICTED: ubiquitin-related modifier 1 homolog 2 [Malus domestica] |
| 8 |
Hb_017259_010 |
0.1039040582 |
- |
- |
- |
| 9 |
Hb_000394_190 |
0.1063292324 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_011848_010 |
0.1068519933 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 11 |
Hb_000261_250 |
0.1080472747 |
- |
- |
elongation factor 1-alpha [Lepidium sativum] |
| 12 |
Hb_012114_060 |
0.1082882274 |
- |
- |
PREDICTED: uncharacterized protein LOC105645814 [Jatropha curcas] |
| 13 |
Hb_000785_030 |
0.1107885618 |
- |
- |
hypothetical protein RCOM_0808030 [Ricinus communis] |
| 14 |
Hb_002530_010 |
0.1135804552 |
- |
- |
hypothetical protein EUGRSUZ_C02580 [Eucalyptus grandis] |
| 15 |
Hb_000787_170 |
0.11416866 |
- |
- |
hypothetical protein POPTR_0005s06400g [Populus trichocarpa] |
| 16 |
Hb_003091_010 |
0.1144021458 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 17 |
Hb_000976_260 |
0.1150381619 |
- |
- |
- |
| 18 |
Hb_000106_130 |
0.1151097943 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_001135_020 |
0.1151895656 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 20 |
Hb_001910_020 |
0.1163611818 |
- |
- |
PREDICTED: ubiquitin-associated domain-containing protein 2 [Jatropha curcas] |