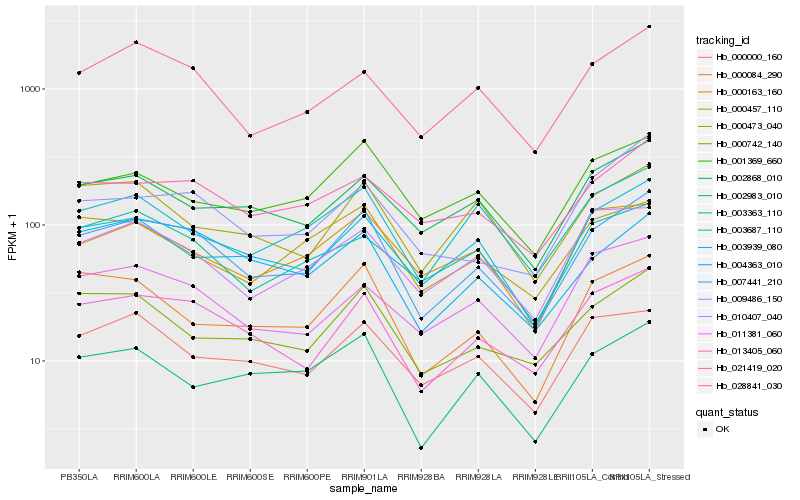
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007441_210 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa013350mg [Prunus persica] |
| 2 |
Hb_013405_060 |
0.0684064831 |
- |
- |
PREDICTED: probable RNA-binding protein 18 isoform X1 [Jatropha curcas] |
| 3 |
Hb_028841_030 |
0.0916343596 |
- |
- |
- |
| 4 |
Hb_004363_010 |
0.0954918755 |
- |
- |
hypothetical protein CISIN_1g0405721mg, partial [Citrus sinensis] |
| 5 |
Hb_021419_020 |
0.0970661287 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 6 |
Hb_000742_140 |
0.0985109052 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000084_290 |
0.1021688682 |
- |
- |
mitochondrial 39S ribosomal protein L53 [Hevea brasiliensis] |
| 8 |
Hb_003939_080 |
0.1051615567 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002868_010 |
0.1055383303 |
- |
- |
60S ribosomal protein L30, putative [Ricinus communis] |
| 10 |
Hb_000000_160 |
0.1076404203 |
- |
- |
PREDICTED: membrin-11 [Jatropha curcas] |
| 11 |
Hb_009486_150 |
0.1096189787 |
- |
- |
60S ribosomal protein L37a-2 [Medicago truncatula] |
| 12 |
Hb_010407_040 |
0.1096708634 |
- |
- |
PREDICTED: uncharacterized protein At4g14342 isoform X1 [Nelumbo nucifera] |
| 13 |
Hb_003363_110 |
0.1101061656 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_001369_660 |
0.1101402443 |
- |
- |
hypothetical protein B456_001G174200, partial [Gossypium raimondii] |
| 15 |
Hb_003687_110 |
0.1105242725 |
- |
- |
PREDICTED: uncharacterized protein At4g14342 isoform X1 [Malus domestica] |
| 16 |
Hb_002983_010 |
0.1105522395 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 17 |
Hb_000473_040 |
0.1116395111 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: RNA polymerase II transcriptional coactivator KIWI-like [Jatropha curcas] |
| 18 |
Hb_000457_110 |
0.1116758175 |
- |
- |
Mitochondrial ribosomal protein L37 isoform 1 [Theobroma cacao] |
| 19 |
Hb_011381_060 |
0.1127073026 |
- |
- |
PREDICTED: ADP-ribosylation factor 3 [Jatropha curcas] |
| 20 |
Hb_000163_160 |
0.1128367028 |
- |
- |
Putative small nuclear ribonucleoprotein Sm D2 [Glycine soja] |