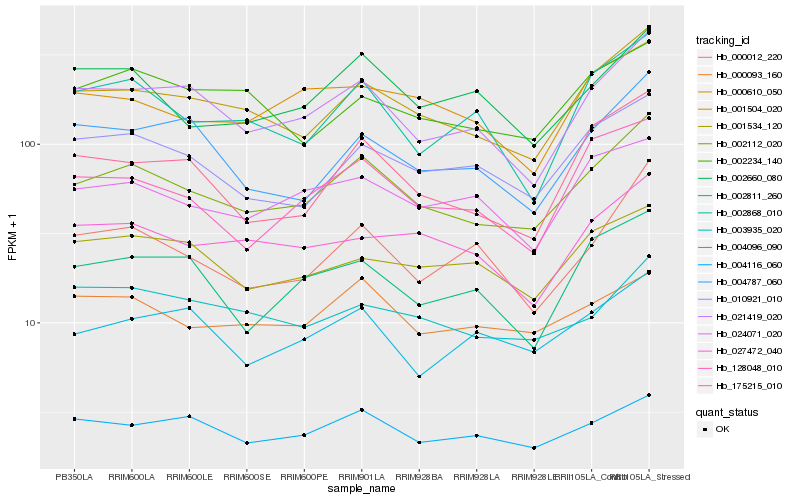
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002112_020 |
0.0 |
- |
- |
60S ribosomal protein L9, putative [Ricinus communis] |
| 2 |
Hb_001504_020 |
0.0552675171 |
- |
- |
40S ribosomal protein S12A [Hevea brasiliensis] |
| 3 |
Hb_021419_020 |
0.0701865502 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 4 |
Hb_010921_010 |
0.0827060725 |
- |
- |
Small nuclear ribonucleoprotein family protein [Theobroma cacao] |
| 5 |
Hb_002660_080 |
0.0866449015 |
- |
- |
60S ribosomal protein L12A [Hevea brasiliensis] |
| 6 |
Hb_128048_010 |
0.0882771182 |
- |
- |
small nuclear ribonucleoprotein U)1a,U)2b, putative [Ricinus communis] |
| 7 |
Hb_175215_010 |
0.0886282495 |
- |
- |
hypothetical protein CISIN_1g029605mg [Citrus sinensis] |
| 8 |
Hb_004116_060 |
0.0894515928 |
- |
- |
PREDICTED: nuclear pore complex protein NUP88 [Jatropha curcas] |
| 9 |
Hb_024071_020 |
0.0911274102 |
- |
- |
PREDICTED: uncharacterized protein LOC105639166 [Jatropha curcas] |
| 10 |
Hb_027472_040 |
0.0915259522 |
- |
- |
SKP1 [Hevea brasiliensis] |
| 11 |
Hb_000093_160 |
0.0926487146 |
- |
- |
PREDICTED: nuclear transcription factor Y subunit B-1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000012_220 |
0.0928567469 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002868_010 |
0.0948033811 |
- |
- |
60S ribosomal protein L30, putative [Ricinus communis] |
| 14 |
Hb_004096_090 |
0.0949445988 |
- |
- |
Plastid-specific 30S ribosomal protein 3, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_002234_140 |
0.0958248796 |
- |
- |
DNA-directed RNA polymerase II family protein [Populus trichocarpa] |
| 16 |
Hb_003935_020 |
0.096139624 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_002811_260 |
0.0963155417 |
- |
- |
PREDICTED: PITH domain-containing protein At3g04780 [Jatropha curcas] |
| 18 |
Hb_000610_050 |
0.0964586885 |
- |
- |
skp1, putative [Ricinus communis] |
| 19 |
Hb_004787_060 |
0.0973268127 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 20 |
Hb_001534_120 |
0.0984691347 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 37 homolog 1 [Gossypium raimondii] |