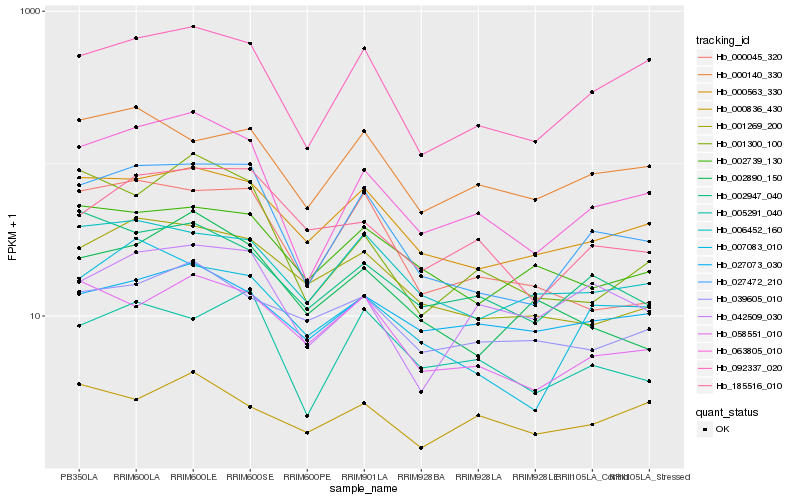
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_063805_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_027472_210 |
0.0949476988 |
- |
- |
PREDICTED: uncharacterized protein LOC105646181 [Jatropha curcas] |
| 3 |
Hb_092337_020 |
0.108513149 |
- |
- |
hypothetical protein B456_013G264900 [Gossypium raimondii] |
| 4 |
Hb_001300_100 |
0.1253353692 |
- |
- |
- |
| 5 |
Hb_000563_330 |
0.1284788282 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_002947_040 |
0.1384557682 |
- |
- |
PREDICTED: uncharacterized protein LOC105131897 isoform X1 [Populus euphratica] |
| 7 |
Hb_042509_030 |
0.1411500496 |
- |
- |
PREDICTED: B-box zinc finger protein 22 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001269_200 |
0.1443786244 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 9 |
Hb_058551_010 |
0.1446463375 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 10 |
Hb_185516_010 |
0.14532134 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 11 |
Hb_007083_010 |
0.1474079267 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 13-like, partial [Nelumbo nucifera] |
| 12 |
Hb_006452_160 |
0.148490801 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 13 |
Hb_027073_030 |
0.1487125189 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 14 |
Hb_005291_040 |
0.1520392049 |
- |
- |
PREDICTED: gamma-gliadin [Jatropha curcas] |
| 15 |
Hb_002739_130 |
0.1526138659 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Populus euphratica] |
| 16 |
Hb_000140_330 |
0.1530761014 |
- |
- |
PREDICTED: uncharacterized protein LOC105642909 [Jatropha curcas] |
| 17 |
Hb_000836_430 |
0.1535075906 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61520, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_002890_150 |
0.154117277 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000045_320 |
0.1542718731 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 20 |
Hb_039605_010 |
0.1547785021 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |