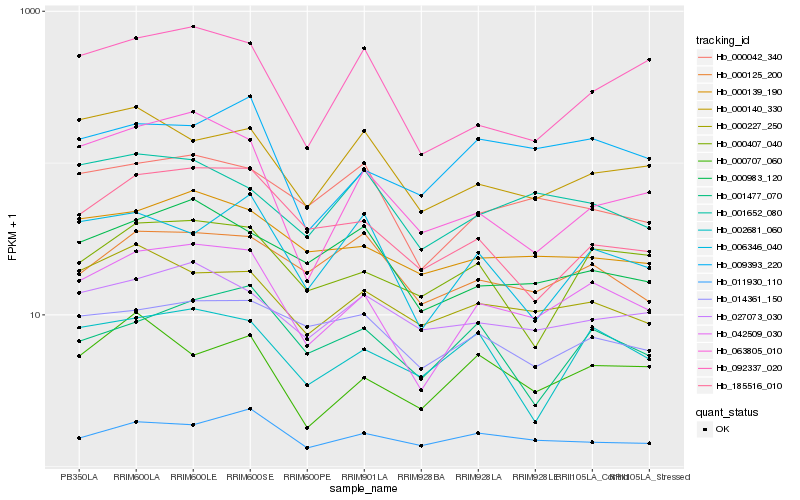
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_042509_030 |
0.0 |
- |
- |
PREDICTED: B-box zinc finger protein 22 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000139_190 |
0.1328493044 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_009393_220 |
0.1329916091 |
transcription factor |
TF Family: Tify |
JAZ11 [Hevea brasiliensis] |
| 4 |
Hb_000227_250 |
0.135146321 |
transcription factor |
TF Family: SNF2 |
PREDICTED: probable ATP-dependent DNA helicase CHR12 [Jatropha curcas] |
| 5 |
Hb_092337_020 |
0.1355903663 |
- |
- |
hypothetical protein B456_013G264900 [Gossypium raimondii] |
| 6 |
Hb_000407_040 |
0.1365097386 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_000042_340 |
0.1371423756 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 8 |
Hb_185516_010 |
0.1383791485 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 9 |
Hb_000983_120 |
0.1389237193 |
- |
- |
cyclin-dependent protein kinase, putative [Ricinus communis] |
| 10 |
Hb_027073_030 |
0.1408000269 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 11 |
Hb_000707_060 |
0.1410385172 |
- |
- |
- |
| 12 |
Hb_063805_010 |
0.1411500496 |
- |
- |
- |
| 13 |
Hb_011930_110 |
0.1443317558 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g59720, mitochondrial [Jatropha curcas] |
| 14 |
Hb_006346_040 |
0.1448311782 |
- |
- |
hypothetical protein JCGZ_20335 [Jatropha curcas] |
| 15 |
Hb_002681_060 |
0.1454056525 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 16 |
Hb_001652_080 |
0.1491162783 |
- |
- |
PREDICTED: uncharacterized protein LOC105643776 [Jatropha curcas] |
| 17 |
Hb_001477_070 |
0.1496224003 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000140_330 |
0.1502018115 |
- |
- |
PREDICTED: uncharacterized protein LOC105642909 [Jatropha curcas] |
| 19 |
Hb_014361_150 |
0.1523714051 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 20 |
Hb_000125_200 |
0.1528894307 |
transcription factor |
TF Family: bZIP |
PREDICTED: probable transcription factor PosF21 [Jatropha curcas] |