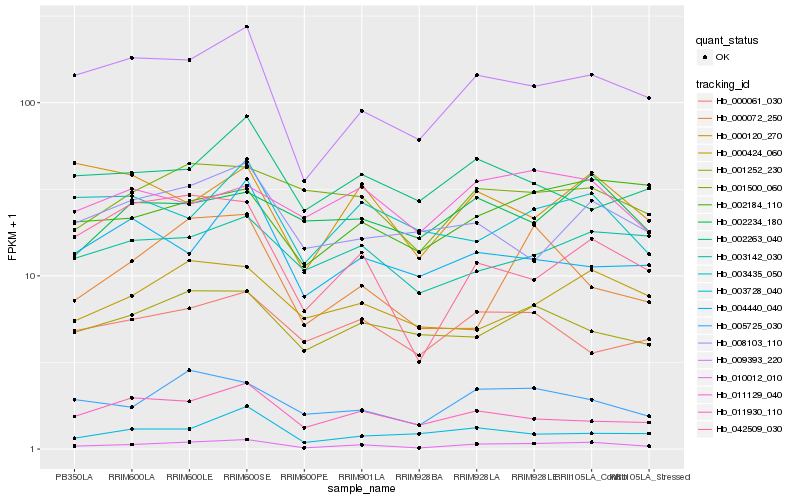
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010012_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_009393_220 |
0.1196031267 |
transcription factor |
TF Family: Tify |
JAZ11 [Hevea brasiliensis] |
| 3 |
Hb_005725_030 |
0.1416746063 |
- |
- |
PREDICTED: U-box domain-containing protein 27-like [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_011930_110 |
0.1611579467 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g59720, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000424_060 |
0.1624057469 |
- |
- |
PREDICTED: myosin-15 [Jatropha curcas] |
| 6 |
Hb_042509_030 |
0.1634213816 |
- |
- |
PREDICTED: B-box zinc finger protein 22 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002184_110 |
0.1636608939 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 [Jatropha curcas] |
| 8 |
Hb_001500_060 |
0.1637405091 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein ZFN-like [Jatropha curcas] |
| 9 |
Hb_001252_230 |
0.1637563511 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003142_030 |
0.1686408351 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 14, chloroplastic [Jatropha curcas] |
| 11 |
Hb_003728_040 |
0.1689015836 |
- |
- |
PREDICTED: uncharacterized protein LOC101492305 [Cicer arietinum] |
| 12 |
Hb_000072_250 |
0.1701278719 |
transcription factor |
TF Family: Orphans |
- |
| 13 |
Hb_003435_050 |
0.1703715062 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1-like [Jatropha curcas] |
| 14 |
Hb_008103_110 |
0.1735987322 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_000061_030 |
0.1762358104 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065 [Jatropha curcas] |
| 16 |
Hb_000120_270 |
0.1767765159 |
- |
- |
PREDICTED: histone deacetylase 8 [Jatropha curcas] |
| 17 |
Hb_011129_040 |
0.1780522086 |
- |
- |
PREDICTED: uncharacterized protein LOC105645602 [Jatropha curcas] |
| 18 |
Hb_002234_180 |
0.1781659401 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004440_040 |
0.1799298936 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_002263_040 |
0.182043044 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |