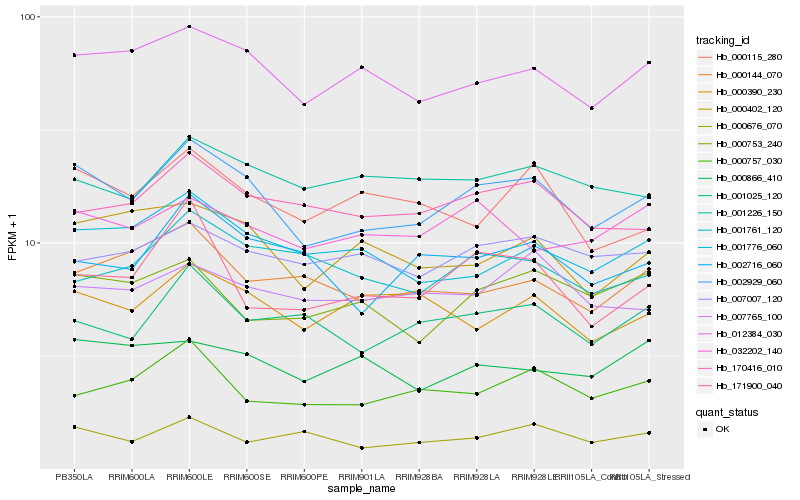
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002929_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_012384_030 |
0.0818492924 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase isozyme L5-like [Jatropha curcas] |
| 3 |
Hb_000402_120 |
0.0969911712 |
- |
- |
PREDICTED: uncharacterized protein LOC105634656 [Jatropha curcas] |
| 4 |
Hb_001776_060 |
0.0982947065 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 5-like [Jatropha curcas] |
| 5 |
Hb_170416_010 |
0.0993160777 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 6 |
Hb_001025_120 |
0.1006546953 |
transcription factor |
TF Family: Jumonji |
PREDICTED: jmjC domain-containing protein 4 [Jatropha curcas] |
| 7 |
Hb_002716_060 |
0.1022138391 |
- |
- |
carboxylesterase np, putative [Ricinus communis] |
| 8 |
Hb_000115_280 |
0.1030250753 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 9 |
Hb_007007_120 |
0.1052600039 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001226_150 |
0.1055490206 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 11 |
Hb_000390_230 |
0.105668956 |
- |
- |
PREDICTED: transducin beta-like protein 3 [Jatropha curcas] |
| 12 |
Hb_032202_140 |
0.1062027885 |
- |
- |
PREDICTED: flap endonuclease GEN-like 1 [Jatropha curcas] |
| 13 |
Hb_171900_040 |
0.1069968636 |
- |
- |
PREDICTED: probable mitochondrial import inner membrane translocase subunit TIM21 [Jatropha curcas] |
| 14 |
Hb_000753_240 |
0.1075044231 |
- |
- |
PREDICTED: ADP,ATP carrier protein ER-ANT1 [Jatropha curcas] |
| 15 |
Hb_007765_100 |
0.107794848 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 16 |
Hb_000144_070 |
0.1088165073 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000866_410 |
0.1089101232 |
- |
- |
hypothetical protein JCGZ_14422 [Jatropha curcas] |
| 18 |
Hb_000676_070 |
0.1089884595 |
- |
- |
PREDICTED: uncharacterized protein LOC105634617 [Jatropha curcas] |
| 19 |
Hb_000757_030 |
0.1093840946 |
- |
- |
radical sam protein, putative [Ricinus communis] |
| 20 |
Hb_001761_120 |
0.1105270128 |
- |
- |
hypothetical protein JCGZ_22564 [Jatropha curcas] |