| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
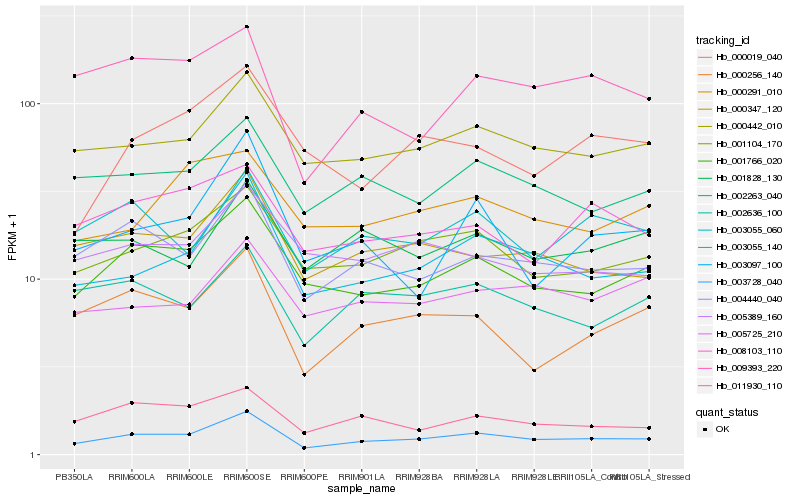
Hb_003728_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC101492305 [Cicer arietinum] |
| 2 |
Hb_003055_140 |
0.080193505 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 3 |
Hb_001766_020 |
0.0913584815 |
- |
- |
PREDICTED: probable galacturonosyltransferase 14 [Populus euphratica] |
| 4 |
Hb_000442_010 |
0.0937841604 |
- |
- |
hypothetical protein POPTR_0010s19060g [Populus trichocarpa] |
| 5 |
Hb_004440_040 |
0.0952945675 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 6 |
Hb_001104_170 |
0.0996597966 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2c [Jatropha curcas] |
| 7 |
Hb_000347_120 |
0.1061059798 |
- |
- |
PREDICTED: uncharacterized protein LOC105631363 [Jatropha curcas] |
| 8 |
Hb_002263_040 |
0.1113676516 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 9 |
Hb_000256_140 |
0.1140893613 |
- |
- |
PREDICTED: uncharacterized protein LOC105637585 isoform X1 [Jatropha curcas] |
| 10 |
Hb_011930_110 |
0.1157519966 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g59720, mitochondrial [Jatropha curcas] |
| 11 |
Hb_003055_060 |
0.1197861381 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105649487 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002636_100 |
0.1234805962 |
- |
- |
PREDICTED: DNA-directed RNA polymerase 1B, mitochondrial [Jatropha curcas] |
| 13 |
Hb_009393_220 |
0.1247508407 |
transcription factor |
TF Family: Tify |
JAZ11 [Hevea brasiliensis] |
| 14 |
Hb_005725_210 |
0.126157799 |
- |
- |
PREDICTED: RNA-binding protein 1 [Jatropha curcas] |
| 15 |
Hb_008103_110 |
0.1280985188 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_005389_160 |
0.1298560454 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 17 |
Hb_003097_100 |
0.1321051385 |
- |
- |
probable glycerol-3-phosphate dehydrogenase [NAD(+)] 1, cytosolic [Jatropha curcas] |
| 18 |
Hb_000019_040 |
0.1329197424 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 3, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000291_010 |
0.1329300025 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 20 |
Hb_001828_130 |
0.1329846229 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |