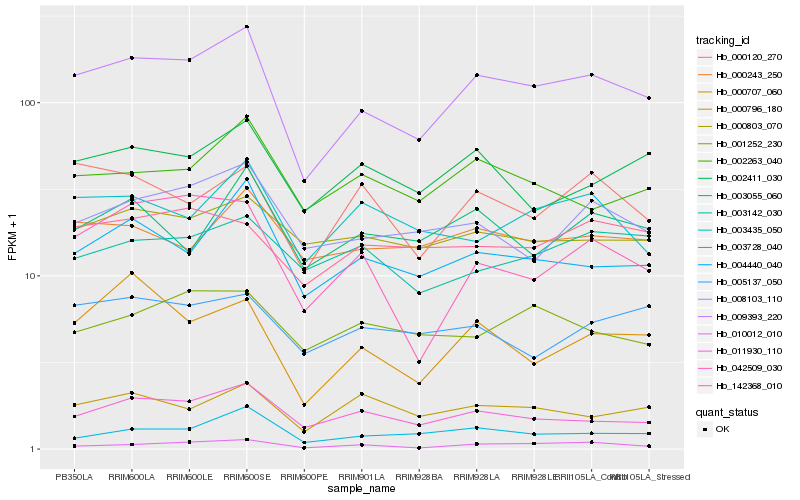
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009393_220 |
0.0 |
transcription factor |
TF Family: Tify |
JAZ11 [Hevea brasiliensis] |
| 2 |
Hb_011930_110 |
0.1036746234 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g59720, mitochondrial [Jatropha curcas] |
| 3 |
Hb_004440_040 |
0.108095772 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_008103_110 |
0.1174300137 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_010012_010 |
0.1196031267 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000120_270 |
0.1243450319 |
- |
- |
PREDICTED: histone deacetylase 8 [Jatropha curcas] |
| 7 |
Hb_003728_040 |
0.1247508407 |
- |
- |
PREDICTED: uncharacterized protein LOC101492305 [Cicer arietinum] |
| 8 |
Hb_002263_040 |
0.1254408946 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 9 |
Hb_002411_030 |
0.1263864479 |
transcription factor |
TF Family: NF-YC |
PREDICTED: dr1-associated corepressor homolog [Jatropha curcas] |
| 10 |
Hb_000243_250 |
0.1288779271 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 44 [Jatropha curcas] |
| 11 |
Hb_000707_060 |
0.129009178 |
- |
- |
- |
| 12 |
Hb_003055_060 |
0.129913341 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105649487 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003435_050 |
0.1307515486 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1-like [Jatropha curcas] |
| 14 |
Hb_000796_180 |
0.1309108016 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Vitis vinifera] |
| 15 |
Hb_000803_070 |
0.1311862972 |
- |
- |
PREDICTED: protein TRAUCO [Jatropha curcas] |
| 16 |
Hb_042509_030 |
0.1329916091 |
- |
- |
PREDICTED: B-box zinc finger protein 22 isoform X1 [Jatropha curcas] |
| 17 |
Hb_142368_010 |
0.1332829837 |
- |
- |
mRNA (guanine-7-)methyltransferase, putative [Ricinus communis] |
| 18 |
Hb_005137_050 |
0.1370251824 |
- |
- |
PREDICTED: nuclear pore complex protein NUP58 [Jatropha curcas] |
| 19 |
Hb_001252_230 |
0.1386299408 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003142_030 |
0.1404214588 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 14, chloroplastic [Jatropha curcas] |