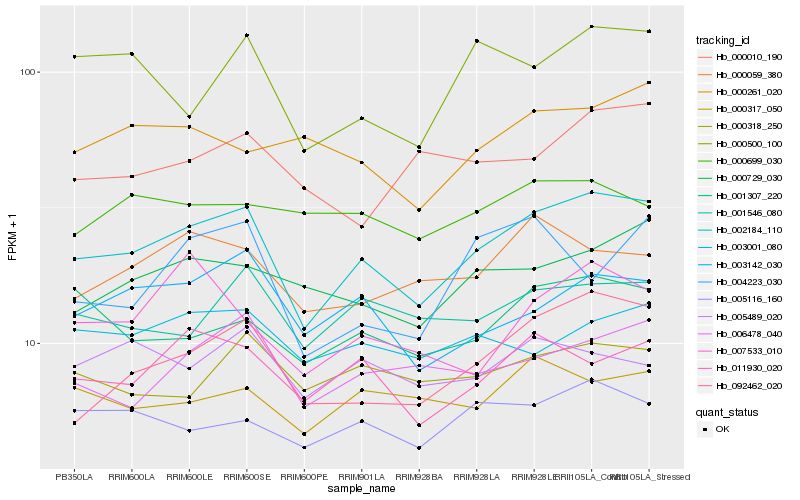
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002184_110 |
0.0 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 [Jatropha curcas] |
| 2 |
Hb_092462_020 |
0.079498023 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_011930_020 |
0.0891100777 |
- |
- |
PREDICTED: uncharacterized protein LOC105649787 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003142_030 |
0.0923921575 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 14, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001307_220 |
0.0934656655 |
- |
- |
PREDICTED: glutathione S-transferase omega-like 2 [Jatropha curcas] |
| 6 |
Hb_006478_040 |
0.0938751109 |
- |
- |
PREDICTED: methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000729_030 |
0.0948779701 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 8 |
Hb_000059_380 |
0.0968605055 |
- |
- |
PREDICTED: uncharacterized protein LOC105648668 [Jatropha curcas] |
| 9 |
Hb_001546_080 |
0.0970979292 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12b isoform X1 [Jatropha curcas] |
| 10 |
Hb_005116_160 |
0.0975611978 |
transcription factor |
TF Family: TRAF |
- |
| 11 |
Hb_000500_100 |
0.1000355609 |
- |
- |
peptide methionine sulfoxide reductase A1-like [Jatropha curcas] |
| 12 |
Hb_003001_080 |
0.1026259237 |
- |
- |
PREDICTED: uncharacterized protein LOC105644363 [Jatropha curcas] |
| 13 |
Hb_000317_050 |
0.1033672303 |
- |
- |
formiminotransferase-cyclodeaminase, putative [Ricinus communis] |
| 14 |
Hb_000318_250 |
0.1047568031 |
- |
- |
PREDICTED: uncharacterized protein LOC105634251 [Jatropha curcas] |
| 15 |
Hb_005489_020 |
0.1062359113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007533_010 |
0.1067389857 |
transcription factor |
TF Family: bZIP |
G-box-binding factor, putative [Ricinus communis] |
| 17 |
Hb_000261_020 |
0.1093249337 |
- |
- |
PREDICTED: chaperone protein dnaJ 20, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000010_190 |
0.1097530154 |
- |
- |
PREDICTED: protein-L-isoaspartate O-methyltransferase 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_004223_030 |
0.1106590554 |
- |
- |
PREDICTED: uncharacterized protein LOC105635347 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000699_030 |
0.1122456675 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1-like isoform X2 [Jatropha curcas] |