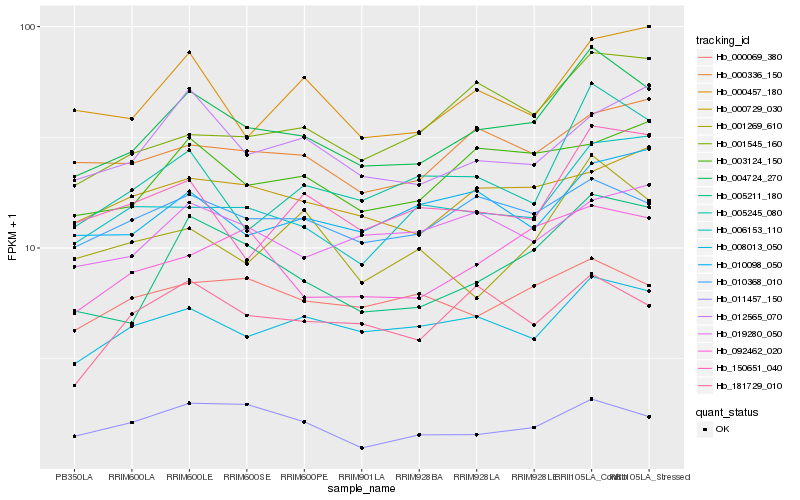
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004724_270 |
0.0 |
- |
- |
hypothetical protein JCGZ_12706 [Jatropha curcas] |
| 2 |
Hb_005211_180 |
0.0801578163 |
- |
- |
Transmembrane protein TPARL, putative [Ricinus communis] |
| 3 |
Hb_008013_050 |
0.0911907926 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At1g33420 [Jatropha curcas] |
| 4 |
Hb_010368_010 |
0.0986533775 |
- |
- |
RNA splicing protein mrs2, mitochondrial, putative [Ricinus communis] |
| 5 |
Hb_005245_080 |
0.0992780676 |
- |
- |
PREDICTED: uncharacterized protein LOC105643645 isoform X2 [Jatropha curcas] |
| 6 |
Hb_092462_020 |
0.1014107048 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_003124_150 |
0.1043733986 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 28-like [Jatropha curcas] |
| 8 |
Hb_000457_180 |
0.1058227386 |
- |
- |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial [Jatropha curcas] |
| 9 |
Hb_006153_110 |
0.108061361 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 10 |
Hb_000336_150 |
0.1086156918 |
- |
- |
ribulose-5-phosphate-3-epimerase, putative [Ricinus communis] |
| 11 |
Hb_001545_160 |
0.1097344743 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC102615208 isoform X1 [Citrus sinensis] |
| 12 |
Hb_150651_040 |
0.10979533 |
- |
- |
PREDICTED: random slug protein 5-like [Jatropha curcas] |
| 13 |
Hb_010098_050 |
0.1113620371 |
- |
- |
PREDICTED: putative deoxyribonuclease tatdn3-A [Jatropha curcas] |
| 14 |
Hb_012565_070 |
0.1114133247 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 15 |
Hb_181729_010 |
0.1116614639 |
- |
- |
PREDICTED: DNA polymerase epsilon subunit 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000729_030 |
0.1120265003 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 17 |
Hb_000069_380 |
0.1124817619 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 18 |
Hb_011457_150 |
0.1132401485 |
- |
- |
PREDICTED: uncharacterized protein LOC105649946 [Jatropha curcas] |
| 19 |
Hb_019280_050 |
0.1145934375 |
- |
- |
PREDICTED: protein SAWADEE HOMEODOMAIN HOMOLOG 2 [Jatropha curcas] |
| 20 |
Hb_001269_610 |
0.1146916878 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |