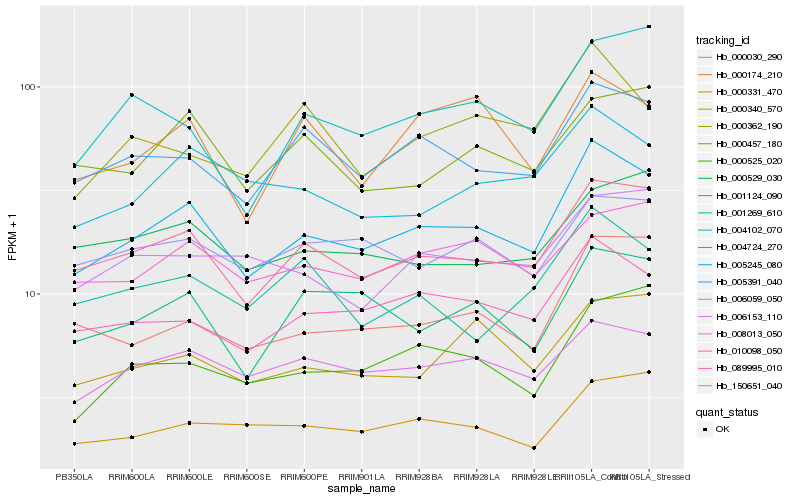
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005245_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643645 isoform X2 [Jatropha curcas] |
| 2 |
Hb_150651_040 |
0.0611881126 |
- |
- |
PREDICTED: random slug protein 5-like [Jatropha curcas] |
| 3 |
Hb_089995_010 |
0.0836322887 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like [Eucalyptus grandis] |
| 4 |
Hb_005391_040 |
0.0874940746 |
- |
- |
PREDICTED: 1,4-dihydroxy-2-naphthoyl-CoA thioesterase 1 [Jatropha curcas] |
| 5 |
Hb_008013_050 |
0.0966733241 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein At1g33420 [Jatropha curcas] |
| 6 |
Hb_000030_290 |
0.0977080892 |
- |
- |
PREDICTED: uncharacterized protein LOC100247068 [Vitis vinifera] |
| 7 |
Hb_004724_270 |
0.0992780676 |
- |
- |
hypothetical protein JCGZ_12706 [Jatropha curcas] |
| 8 |
Hb_001124_090 |
0.0993570839 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 9 |
Hb_010098_050 |
0.1017224027 |
- |
- |
PREDICTED: putative deoxyribonuclease tatdn3-A [Jatropha curcas] |
| 10 |
Hb_000331_470 |
0.1037602193 |
- |
- |
PREDICTED: uncharacterized protein LOC105136689 [Populus euphratica] |
| 11 |
Hb_000525_020 |
0.1066996605 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_006059_050 |
0.1069684344 |
- |
- |
PREDICTED: polyadenylate-binding protein 1 [Jatropha curcas] |
| 13 |
Hb_000340_570 |
0.1073596834 |
- |
- |
PREDICTED: RNA-binding protein BRN1 [Jatropha curcas] |
| 14 |
Hb_006153_110 |
0.1090300306 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 15 |
Hb_000457_180 |
0.1098891 |
- |
- |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000529_030 |
0.1121523147 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_000174_210 |
0.1133413194 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 18 |
Hb_004102_070 |
0.1135968924 |
- |
- |
Cytochrome c oxidase subunit 5b-2, mitochondrial -like protein [Gossypium arboreum] |
| 19 |
Hb_000362_190 |
0.1144963214 |
- |
- |
PREDICTED: 28 kDa heat- and acid-stable phosphoprotein [Jatropha curcas] |
| 20 |
Hb_001269_610 |
0.1157133447 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |